
Natranaerobius trueperi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Natranaerobiales; Natranaerobiaceae; Natranaerobius
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
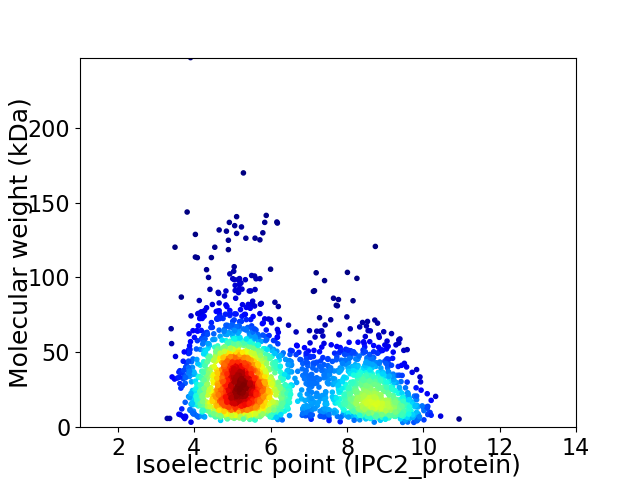
Virtual 2D-PAGE plot for 2464 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A226BXL2|A0A226BXL2_9FIRM Group II intron reverse transcriptase/maturase (Fragment) OS=Natranaerobius trueperi OX=759412 GN=CDO51_11720 PE=4 SV=1
MM1 pKa = 7.43KK2 pKa = 9.17RR3 pKa = 11.84TSLYY7 pKa = 10.76LVLSLFLVIAGAMAVTGCDD26 pKa = 2.98EE27 pKa = 4.24SEE29 pKa = 4.43RR30 pKa = 11.84YY31 pKa = 9.77AIATGGSGGVYY42 pKa = 9.95YY43 pKa = 10.12PLGTAMSNILNDD55 pKa = 3.51EE56 pKa = 4.33LEE58 pKa = 4.46GVDD61 pKa = 4.69ISAEE65 pKa = 4.08STGASVEE72 pKa = 4.26NVNLTANEE80 pKa = 4.17DD81 pKa = 3.4VHH83 pKa = 7.51LALVQNDD90 pKa = 3.01ISYY93 pKa = 10.26YY94 pKa = 8.7AYY96 pKa = 10.23EE97 pKa = 4.82GIEE100 pKa = 4.06MFDD103 pKa = 3.91DD104 pKa = 4.14EE105 pKa = 5.3EE106 pKa = 4.99PMEE109 pKa = 4.43EE110 pKa = 3.9LRR112 pKa = 11.84GMGTLYY118 pKa = 10.39PEE120 pKa = 5.25TIQIVADD127 pKa = 3.67AGEE130 pKa = 4.8DD131 pKa = 3.45IEE133 pKa = 4.86TVEE136 pKa = 4.7DD137 pKa = 3.97LDD139 pKa = 4.06GKK141 pKa = 10.23RR142 pKa = 11.84VAVGAPGSGTEE153 pKa = 3.86ANARR157 pKa = 11.84QLLNAHH163 pKa = 7.48GITYY167 pKa = 10.75DD168 pKa = 5.16DD169 pKa = 4.3IEE171 pKa = 6.07ADD173 pKa = 3.51YY174 pKa = 11.4LSFSEE179 pKa = 5.51ASDD182 pKa = 3.6NLRR185 pKa = 11.84DD186 pKa = 3.7GHH188 pKa = 7.99LDD190 pKa = 3.32AAFVTAGTPTASITDD205 pKa = 4.14LSTTHH210 pKa = 6.72NIDD213 pKa = 3.19LVSISEE219 pKa = 4.11EE220 pKa = 3.68MSEE223 pKa = 4.89EE224 pKa = 3.87IQEE227 pKa = 4.33DD228 pKa = 4.32YY229 pKa = 10.76PYY231 pKa = 10.11YY232 pKa = 11.07AEE234 pKa = 4.06VTIDD238 pKa = 3.13EE239 pKa = 4.66GTYY242 pKa = 10.61SDD244 pKa = 3.99DD245 pKa = 4.03QEE247 pKa = 4.39DD248 pKa = 3.91VRR250 pKa = 11.84ALAVMAMIIVHH261 pKa = 6.95EE262 pKa = 5.05DD263 pKa = 3.18LPEE266 pKa = 5.94DD267 pKa = 3.32IVYY270 pKa = 10.71EE271 pKa = 4.03MTKK274 pKa = 10.78GIFTSTDD281 pKa = 3.17EE282 pKa = 4.57LGEE285 pKa = 3.92AHH287 pKa = 6.17EE288 pKa = 4.93RR289 pKa = 11.84GYY291 pKa = 11.24EE292 pKa = 4.07VTAEE296 pKa = 4.0DD297 pKa = 4.15ALEE300 pKa = 4.32GMSLEE305 pKa = 4.12LHH307 pKa = 6.66LGAEE311 pKa = 4.32RR312 pKa = 11.84FFDD315 pKa = 3.84EE316 pKa = 4.59QQ317 pKa = 3.43
MM1 pKa = 7.43KK2 pKa = 9.17RR3 pKa = 11.84TSLYY7 pKa = 10.76LVLSLFLVIAGAMAVTGCDD26 pKa = 2.98EE27 pKa = 4.24SEE29 pKa = 4.43RR30 pKa = 11.84YY31 pKa = 9.77AIATGGSGGVYY42 pKa = 9.95YY43 pKa = 10.12PLGTAMSNILNDD55 pKa = 3.51EE56 pKa = 4.33LEE58 pKa = 4.46GVDD61 pKa = 4.69ISAEE65 pKa = 4.08STGASVEE72 pKa = 4.26NVNLTANEE80 pKa = 4.17DD81 pKa = 3.4VHH83 pKa = 7.51LALVQNDD90 pKa = 3.01ISYY93 pKa = 10.26YY94 pKa = 8.7AYY96 pKa = 10.23EE97 pKa = 4.82GIEE100 pKa = 4.06MFDD103 pKa = 3.91DD104 pKa = 4.14EE105 pKa = 5.3EE106 pKa = 4.99PMEE109 pKa = 4.43EE110 pKa = 3.9LRR112 pKa = 11.84GMGTLYY118 pKa = 10.39PEE120 pKa = 5.25TIQIVADD127 pKa = 3.67AGEE130 pKa = 4.8DD131 pKa = 3.45IEE133 pKa = 4.86TVEE136 pKa = 4.7DD137 pKa = 3.97LDD139 pKa = 4.06GKK141 pKa = 10.23RR142 pKa = 11.84VAVGAPGSGTEE153 pKa = 3.86ANARR157 pKa = 11.84QLLNAHH163 pKa = 7.48GITYY167 pKa = 10.75DD168 pKa = 5.16DD169 pKa = 4.3IEE171 pKa = 6.07ADD173 pKa = 3.51YY174 pKa = 11.4LSFSEE179 pKa = 5.51ASDD182 pKa = 3.6NLRR185 pKa = 11.84DD186 pKa = 3.7GHH188 pKa = 7.99LDD190 pKa = 3.32AAFVTAGTPTASITDD205 pKa = 4.14LSTTHH210 pKa = 6.72NIDD213 pKa = 3.19LVSISEE219 pKa = 4.11EE220 pKa = 3.68MSEE223 pKa = 4.89EE224 pKa = 3.87IQEE227 pKa = 4.33DD228 pKa = 4.32YY229 pKa = 10.76PYY231 pKa = 10.11YY232 pKa = 11.07AEE234 pKa = 4.06VTIDD238 pKa = 3.13EE239 pKa = 4.66GTYY242 pKa = 10.61SDD244 pKa = 3.99DD245 pKa = 4.03QEE247 pKa = 4.39DD248 pKa = 3.91VRR250 pKa = 11.84ALAVMAMIIVHH261 pKa = 6.95EE262 pKa = 5.05DD263 pKa = 3.18LPEE266 pKa = 5.94DD267 pKa = 3.32IVYY270 pKa = 10.71EE271 pKa = 4.03MTKK274 pKa = 10.78GIFTSTDD281 pKa = 3.17EE282 pKa = 4.57LGEE285 pKa = 3.92AHH287 pKa = 6.17EE288 pKa = 4.93RR289 pKa = 11.84GYY291 pKa = 11.24EE292 pKa = 4.07VTAEE296 pKa = 4.0DD297 pKa = 4.15ALEE300 pKa = 4.32GMSLEE305 pKa = 4.12LHH307 pKa = 6.66LGAEE311 pKa = 4.32RR312 pKa = 11.84FFDD315 pKa = 3.84EE316 pKa = 4.59QQ317 pKa = 3.43
Molecular weight: 34.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A226BWZ8|A0A226BWZ8_9FIRM Peptidase_M23 domain-containing protein OS=Natranaerobius trueperi OX=759412 GN=CDO51_08275 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.69KK9 pKa = 7.88RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.23TKK25 pKa = 10.17AGRR28 pKa = 11.84NILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.15GRR39 pKa = 11.84KK40 pKa = 9.03SLTAA44 pKa = 4.07
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.69KK9 pKa = 7.88RR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.39KK14 pKa = 8.49EE15 pKa = 3.5HH16 pKa = 6.11GFRR19 pKa = 11.84KK20 pKa = 9.77RR21 pKa = 11.84MKK23 pKa = 9.23TKK25 pKa = 10.17AGRR28 pKa = 11.84NILRR32 pKa = 11.84NRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.15GRR39 pKa = 11.84KK40 pKa = 9.03SLTAA44 pKa = 4.07
Molecular weight: 5.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
713096 |
26 |
2179 |
289.4 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.588 ± 0.05 | 0.882 ± 0.018 |
5.861 ± 0.053 | 8.2 ± 0.068 |
4.225 ± 0.042 | 6.742 ± 0.053 |
1.73 ± 0.02 | 8.411 ± 0.045 |
7.688 ± 0.057 | 9.906 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.334 ± 0.023 | 5.217 ± 0.038 |
3.452 ± 0.027 | 3.203 ± 0.031 |
3.84 ± 0.036 | 6.234 ± 0.038 |
5.292 ± 0.033 | 6.864 ± 0.042 |
0.742 ± 0.017 | 3.592 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
