
Kwoniella dejecticola CBS 10117
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Kwoniella; Kwoniella dejecticola
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
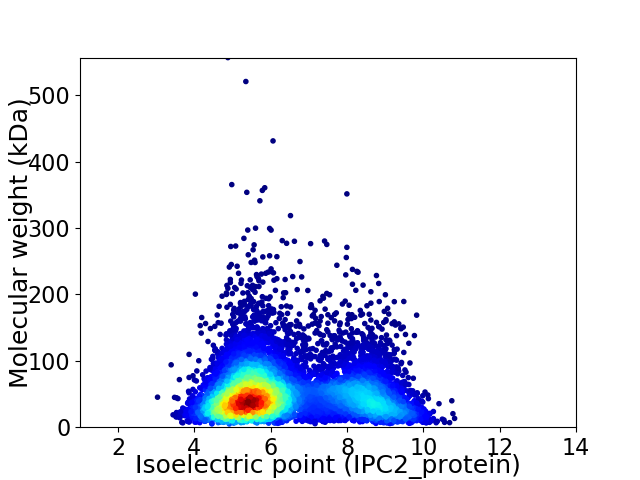
Virtual 2D-PAGE plot for 8600 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A5ZXR9|A0A1A5ZXR9_9TREE Uncharacterized protein OS=Kwoniella dejecticola CBS 10117 OX=1296121 GN=I303_07364 PE=4 SV=1
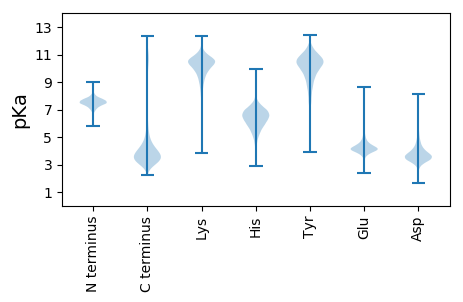
MM1 pKa = 7.72KK2 pKa = 10.21PRR4 pKa = 11.84DD5 pKa = 3.91SPVADD10 pKa = 3.61DD11 pKa = 3.68TFGGIEE17 pKa = 4.11VFDD20 pKa = 4.14AAGDD24 pKa = 3.83SMGYY28 pKa = 9.39ISKK31 pKa = 10.35NLDD34 pKa = 3.06EE35 pKa = 5.18GGYY38 pKa = 9.15TITTDD43 pKa = 3.09PTQAVSISFTQDD55 pKa = 2.08GSNPFSITISSNDD68 pKa = 3.25RR69 pKa = 11.84QAGYY73 pKa = 9.06PYY75 pKa = 10.86LGATLNTGSDD85 pKa = 3.44MGVSSAAFANLGGISHH101 pKa = 7.65IISGPSEE108 pKa = 4.11SDD110 pKa = 3.12DD111 pKa = 3.33TGEE114 pKa = 4.2SNTRR118 pKa = 11.84QLYY121 pKa = 10.37AGSEE125 pKa = 4.25TAIFLYY131 pKa = 10.87NADD134 pKa = 3.73TNAITGQWTNTDD146 pKa = 3.1NTKK149 pKa = 10.39VDD151 pKa = 3.84TIFYY155 pKa = 9.33FGPKK159 pKa = 9.64DD160 pKa = 3.67SYY162 pKa = 10.83SLGMIAPEE170 pKa = 4.73NYY172 pKa = 9.91DD173 pKa = 2.97QFAIDD178 pKa = 4.81FPEE181 pKa = 4.48DD182 pKa = 3.49QQKK185 pKa = 7.81VTFKK189 pKa = 10.65YY190 pKa = 10.96VSIDD194 pKa = 3.35PTPGVV199 pKa = 3.54
MM1 pKa = 7.72KK2 pKa = 10.21PRR4 pKa = 11.84DD5 pKa = 3.91SPVADD10 pKa = 3.61DD11 pKa = 3.68TFGGIEE17 pKa = 4.11VFDD20 pKa = 4.14AAGDD24 pKa = 3.83SMGYY28 pKa = 9.39ISKK31 pKa = 10.35NLDD34 pKa = 3.06EE35 pKa = 5.18GGYY38 pKa = 9.15TITTDD43 pKa = 3.09PTQAVSISFTQDD55 pKa = 2.08GSNPFSITISSNDD68 pKa = 3.25RR69 pKa = 11.84QAGYY73 pKa = 9.06PYY75 pKa = 10.86LGATLNTGSDD85 pKa = 3.44MGVSSAAFANLGGISHH101 pKa = 7.65IISGPSEE108 pKa = 4.11SDD110 pKa = 3.12DD111 pKa = 3.33TGEE114 pKa = 4.2SNTRR118 pKa = 11.84QLYY121 pKa = 10.37AGSEE125 pKa = 4.25TAIFLYY131 pKa = 10.87NADD134 pKa = 3.73TNAITGQWTNTDD146 pKa = 3.1NTKK149 pKa = 10.39VDD151 pKa = 3.84TIFYY155 pKa = 9.33FGPKK159 pKa = 9.64DD160 pKa = 3.67SYY162 pKa = 10.83SLGMIAPEE170 pKa = 4.73NYY172 pKa = 9.91DD173 pKa = 2.97QFAIDD178 pKa = 4.81FPEE181 pKa = 4.48DD182 pKa = 3.49QQKK185 pKa = 7.81VTFKK189 pKa = 10.65YY190 pKa = 10.96VSIDD194 pKa = 3.35PTPGVV199 pKa = 3.54
Molecular weight: 21.25 kDa
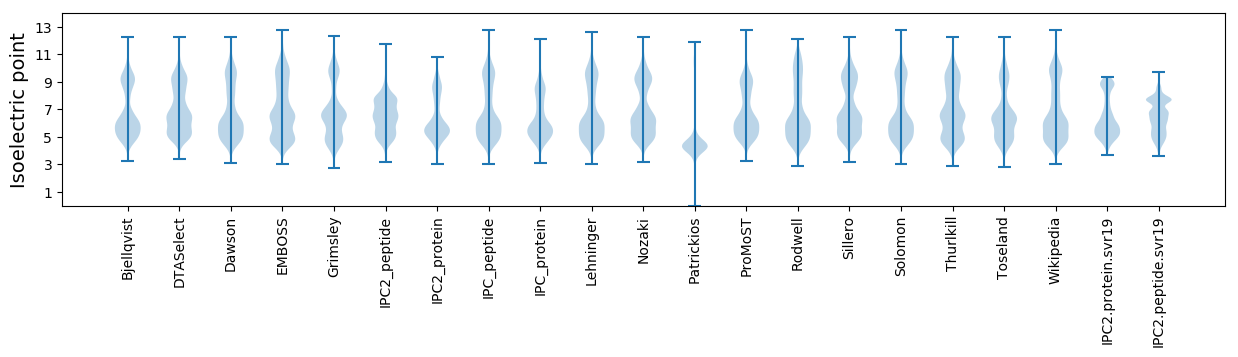
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A5ZY65|A0A1A5ZY65_9TREE Multidrug transporter OS=Kwoniella dejecticola CBS 10117 OX=1296121 GN=I303_07516 PE=4 SV=1
MM1 pKa = 7.77SDD3 pKa = 2.61SGYY6 pKa = 10.87QVMALSPEE14 pKa = 4.36TSPQTPNITPSPPLRR29 pKa = 11.84QLSMSVPLALSADD42 pKa = 3.76GSSAIHH48 pKa = 6.8ASPVPPPRR56 pKa = 11.84DD57 pKa = 3.07IRR59 pKa = 11.84ALLSHH64 pKa = 6.89SPPLTPRR71 pKa = 11.84SCKK74 pKa = 10.22SSSTSPDD81 pKa = 3.04LKK83 pKa = 11.09KK84 pKa = 10.46LDD86 pKa = 3.88KK87 pKa = 10.81RR88 pKa = 11.84STASPSNFSRR98 pKa = 11.84NLPPLTRR105 pKa = 11.84TPPILPRR112 pKa = 11.84PPIGLSLAGPSGNGRR127 pKa = 11.84GTGIGLDD134 pKa = 3.66AFRR137 pKa = 11.84LGSPPKK143 pKa = 10.0QHH145 pKa = 6.51PQPGTATSHH154 pKa = 5.14HH155 pKa = 6.17HH156 pKa = 5.96TRR158 pKa = 11.84SFWVPSSTRR167 pKa = 11.84NDD169 pKa = 2.98QRR171 pKa = 11.84SQHH174 pKa = 5.37QRR176 pKa = 11.84LASLPSIHH184 pKa = 6.57TGHH187 pKa = 6.97RR188 pKa = 11.84EE189 pKa = 3.6KK190 pKa = 10.24TVYY193 pKa = 8.37RR194 pKa = 11.84TPVCIASQRR203 pKa = 11.84SFPMSTLTPVTQGEE217 pKa = 4.18ISSTISGRR225 pKa = 11.84EE226 pKa = 3.8SAYY229 pKa = 10.18QPPPHH234 pKa = 6.84HH235 pKa = 6.75RR236 pKa = 11.84HH237 pKa = 5.08QSLSYY242 pKa = 10.45SSHH245 pKa = 5.94TPLFQDD251 pKa = 3.31EE252 pKa = 4.66PVTHH256 pKa = 6.86RR257 pKa = 11.84RR258 pKa = 11.84HH259 pKa = 6.11PSSGDD264 pKa = 3.14AVIRR268 pKa = 11.84FASHH272 pKa = 6.81PLHH275 pKa = 6.45RR276 pKa = 11.84PAWAAPPPPPAHH288 pKa = 7.02AISIPGRR295 pKa = 11.84TQTMARR301 pKa = 11.84PRR303 pKa = 11.84LQIHH307 pKa = 7.12PYY309 pKa = 9.78APPRR313 pKa = 11.84MDD315 pKa = 3.05PRR317 pKa = 11.84YY318 pKa = 9.78YY319 pKa = 10.5SAGGLGMQTHH329 pKa = 5.96THH331 pKa = 6.98PGLAHH336 pKa = 6.78GGHH339 pKa = 6.36ILGYY343 pKa = 9.61GRR345 pKa = 11.84EE346 pKa = 4.13IVFTSPSSGISPGSFKK362 pKa = 10.93APRR365 pKa = 11.84KK366 pKa = 9.75RR367 pKa = 11.84ADD369 pKa = 3.57DD370 pKa = 3.7SQLAILNDD378 pKa = 3.72VFEE381 pKa = 4.43KK382 pKa = 9.93TAYY385 pKa = 9.72PSTDD389 pKa = 3.02EE390 pKa = 4.2RR391 pKa = 11.84DD392 pKa = 3.29EE393 pKa = 4.18LARR396 pKa = 11.84KK397 pKa = 9.91LGMTSRR403 pKa = 11.84SVQIWFQNRR412 pKa = 11.84RR413 pKa = 11.84RR414 pKa = 11.84AVKK417 pKa = 10.22VDD419 pKa = 3.18AQSAVQRR426 pKa = 11.84AEE428 pKa = 4.31AEE430 pKa = 3.88VDD432 pKa = 3.45TQLMIRR438 pKa = 11.84GPAPNIPRR446 pKa = 11.84PYY448 pKa = 9.22PVHH451 pKa = 5.37TRR453 pKa = 11.84HH454 pKa = 6.39RR455 pKa = 11.84SEE457 pKa = 4.16SQEE460 pKa = 4.03EE461 pKa = 4.71VPLQPIVSAIDD472 pKa = 3.56LSKK475 pKa = 10.99EE476 pKa = 3.69RR477 pKa = 11.84VFALFPTTRR486 pKa = 11.84PPLVASQSDD495 pKa = 3.66MVMVKK500 pKa = 10.28SEE502 pKa = 3.89VMTPP506 pKa = 3.18
MM1 pKa = 7.77SDD3 pKa = 2.61SGYY6 pKa = 10.87QVMALSPEE14 pKa = 4.36TSPQTPNITPSPPLRR29 pKa = 11.84QLSMSVPLALSADD42 pKa = 3.76GSSAIHH48 pKa = 6.8ASPVPPPRR56 pKa = 11.84DD57 pKa = 3.07IRR59 pKa = 11.84ALLSHH64 pKa = 6.89SPPLTPRR71 pKa = 11.84SCKK74 pKa = 10.22SSSTSPDD81 pKa = 3.04LKK83 pKa = 11.09KK84 pKa = 10.46LDD86 pKa = 3.88KK87 pKa = 10.81RR88 pKa = 11.84STASPSNFSRR98 pKa = 11.84NLPPLTRR105 pKa = 11.84TPPILPRR112 pKa = 11.84PPIGLSLAGPSGNGRR127 pKa = 11.84GTGIGLDD134 pKa = 3.66AFRR137 pKa = 11.84LGSPPKK143 pKa = 10.0QHH145 pKa = 6.51PQPGTATSHH154 pKa = 5.14HH155 pKa = 6.17HH156 pKa = 5.96TRR158 pKa = 11.84SFWVPSSTRR167 pKa = 11.84NDD169 pKa = 2.98QRR171 pKa = 11.84SQHH174 pKa = 5.37QRR176 pKa = 11.84LASLPSIHH184 pKa = 6.57TGHH187 pKa = 6.97RR188 pKa = 11.84EE189 pKa = 3.6KK190 pKa = 10.24TVYY193 pKa = 8.37RR194 pKa = 11.84TPVCIASQRR203 pKa = 11.84SFPMSTLTPVTQGEE217 pKa = 4.18ISSTISGRR225 pKa = 11.84EE226 pKa = 3.8SAYY229 pKa = 10.18QPPPHH234 pKa = 6.84HH235 pKa = 6.75RR236 pKa = 11.84HH237 pKa = 5.08QSLSYY242 pKa = 10.45SSHH245 pKa = 5.94TPLFQDD251 pKa = 3.31EE252 pKa = 4.66PVTHH256 pKa = 6.86RR257 pKa = 11.84RR258 pKa = 11.84HH259 pKa = 6.11PSSGDD264 pKa = 3.14AVIRR268 pKa = 11.84FASHH272 pKa = 6.81PLHH275 pKa = 6.45RR276 pKa = 11.84PAWAAPPPPPAHH288 pKa = 7.02AISIPGRR295 pKa = 11.84TQTMARR301 pKa = 11.84PRR303 pKa = 11.84LQIHH307 pKa = 7.12PYY309 pKa = 9.78APPRR313 pKa = 11.84MDD315 pKa = 3.05PRR317 pKa = 11.84YY318 pKa = 9.78YY319 pKa = 10.5SAGGLGMQTHH329 pKa = 5.96THH331 pKa = 6.98PGLAHH336 pKa = 6.78GGHH339 pKa = 6.36ILGYY343 pKa = 9.61GRR345 pKa = 11.84EE346 pKa = 4.13IVFTSPSSGISPGSFKK362 pKa = 10.93APRR365 pKa = 11.84KK366 pKa = 9.75RR367 pKa = 11.84ADD369 pKa = 3.57DD370 pKa = 3.7SQLAILNDD378 pKa = 3.72VFEE381 pKa = 4.43KK382 pKa = 9.93TAYY385 pKa = 9.72PSTDD389 pKa = 3.02EE390 pKa = 4.2RR391 pKa = 11.84DD392 pKa = 3.29EE393 pKa = 4.18LARR396 pKa = 11.84KK397 pKa = 9.91LGMTSRR403 pKa = 11.84SVQIWFQNRR412 pKa = 11.84RR413 pKa = 11.84RR414 pKa = 11.84AVKK417 pKa = 10.22VDD419 pKa = 3.18AQSAVQRR426 pKa = 11.84AEE428 pKa = 4.31AEE430 pKa = 3.88VDD432 pKa = 3.45TQLMIRR438 pKa = 11.84GPAPNIPRR446 pKa = 11.84PYY448 pKa = 9.22PVHH451 pKa = 5.37TRR453 pKa = 11.84HH454 pKa = 6.39RR455 pKa = 11.84SEE457 pKa = 4.16SQEE460 pKa = 4.03EE461 pKa = 4.71VPLQPIVSAIDD472 pKa = 3.56LSKK475 pKa = 10.99EE476 pKa = 3.69RR477 pKa = 11.84VFALFPTTRR486 pKa = 11.84PPLVASQSDD495 pKa = 3.66MVMVKK500 pKa = 10.28SEE502 pKa = 3.89VMTPP506 pKa = 3.18
Molecular weight: 55.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4400537 |
44 |
5001 |
511.7 |
56.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.009 ± 0.026 | 0.968 ± 0.008 |
5.651 ± 0.017 | 6.294 ± 0.025 |
3.301 ± 0.015 | 7.174 ± 0.025 |
2.359 ± 0.011 | 5.097 ± 0.018 |
5.164 ± 0.026 | 8.655 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.009 | 3.87 ± 0.015 |
6.529 ± 0.03 | 4.029 ± 0.023 |
5.731 ± 0.024 | 9.512 ± 0.035 |
6.088 ± 0.018 | 5.601 ± 0.019 |
1.345 ± 0.008 | 2.59 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
