
Exophiala sideris
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Exophiala
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
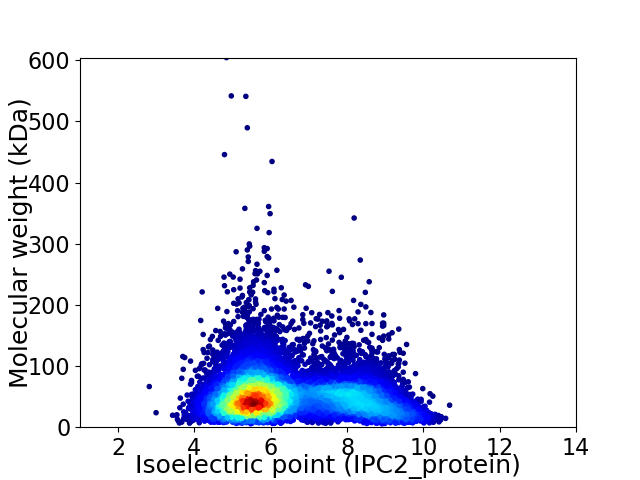
Virtual 2D-PAGE plot for 10841 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D1W667|A0A0D1W667_9EURO Methyltransf_11 domain-containing protein OS=Exophiala sideris OX=1016849 GN=PV11_06125 PE=4 SV=1
MM1 pKa = 7.55KK2 pKa = 10.21SVSSLSITLLLGLATGSVGLPATTEE27 pKa = 4.56DD28 pKa = 4.17GDD30 pKa = 3.95SHH32 pKa = 6.74GWYY35 pKa = 9.78DD36 pKa = 4.01WNPSTAVTSTSTSFSLSVPSSLPGFAGAPASGFGNTNSNGFAPSVSIKK84 pKa = 9.28TVEE87 pKa = 4.07VTKK90 pKa = 10.86TEE92 pKa = 4.16AVCAATEE99 pKa = 4.01EE100 pKa = 4.79SVVGVGTSTGGSDD113 pKa = 3.47SSHH116 pKa = 6.56SGSGSDD122 pKa = 3.34SSSGSGSGSGSGFVSSSGGSWSEE145 pKa = 4.1SGSTYY150 pKa = 10.31YY151 pKa = 10.95AQAPGANSSLSSPTYY166 pKa = 8.86QPYY169 pKa = 9.86VAGPTLLPAVHH180 pKa = 7.14WDD182 pKa = 3.62YY183 pKa = 11.35PVDD186 pKa = 4.73DD187 pKa = 4.79IRR189 pKa = 11.84NLGPINSSDD198 pKa = 4.4LYY200 pKa = 9.15YY201 pKa = 10.38TSNGIADD208 pKa = 4.15PGVQHH213 pKa = 6.38VFASLSTTLQYY224 pKa = 11.31DD225 pKa = 3.91AVVLDD230 pKa = 3.68HH231 pKa = 7.07SSYY234 pKa = 8.97VTSVSCSSTGILVAFEE250 pKa = 4.39SIEE253 pKa = 3.99AFRR256 pKa = 11.84YY257 pKa = 10.36ACDD260 pKa = 2.75SWSAVEE266 pKa = 4.4EE267 pKa = 4.44FVLVTYY273 pKa = 10.52TDD275 pKa = 3.43GCGASDD281 pKa = 4.01DD282 pKa = 4.64QRR284 pKa = 11.84TFWLSNSVEE293 pKa = 4.21ILNNTQSIMVAVEE306 pKa = 3.95TEE308 pKa = 4.05LAIEE312 pKa = 4.01DD313 pKa = 4.59AIYY316 pKa = 10.23GVDD319 pKa = 3.83MVWGNYY325 pKa = 8.96YY326 pKa = 8.44PTGSNSSTSISGSGSGSSSNSTAPSGSGSNSTGLSGSGTNSTVGVNGTTSSNSTSNANGGSSCGAAPSSTIDD398 pKa = 3.41GFPAVACGVSDD409 pKa = 5.89FDD411 pKa = 3.99TQLDD415 pKa = 4.05DD416 pKa = 3.48VLGYY420 pKa = 11.0LDD422 pKa = 5.8FSSTDD427 pKa = 3.23YY428 pKa = 11.22DD429 pKa = 3.81ASLEE433 pKa = 4.09DD434 pKa = 4.27FSPDD438 pKa = 3.03VSFDD442 pKa = 4.5AEE444 pKa = 5.08DD445 pKa = 5.4LDD447 pKa = 6.5DD448 pKa = 6.62DD449 pKa = 4.61DD450 pKa = 4.7ATMEE454 pKa = 3.98RR455 pKa = 11.84RR456 pKa = 11.84ALLGRR461 pKa = 11.84SHH463 pKa = 6.53LRR465 pKa = 11.84KK466 pKa = 9.87RR467 pKa = 11.84GIFSALLSVGEE478 pKa = 4.1QLSSAVVAVATTGVALARR496 pKa = 11.84AAVQVIPGVSNFIASATEE514 pKa = 3.69FDD516 pKa = 4.2PAVHH520 pKa = 6.04GTATLALAPDD530 pKa = 4.85DD531 pKa = 4.54SATSPWGAAAQLYY544 pKa = 7.79TSSSASATTAADD556 pKa = 2.96ITLYY560 pKa = 10.68CVNCGLKK567 pKa = 10.59GHH569 pKa = 5.7VTLSGSAKK577 pKa = 10.2FNVIDD582 pKa = 4.36GLHH585 pKa = 6.18GLSTSINADD594 pKa = 3.36LEE596 pKa = 4.61AGLNLGLVAQATFKK610 pKa = 10.04DD611 pKa = 4.03TKK613 pKa = 9.51TKK615 pKa = 11.0SLIKK619 pKa = 10.26QAIPDD624 pKa = 3.8LGVSVKK630 pKa = 10.5GVFAAGVYY638 pKa = 9.81IAVDD642 pKa = 3.78AVATLDD648 pKa = 4.89IEE650 pKa = 4.56AQGQALIGITMTISDD665 pKa = 4.37FQATLNLLDD674 pKa = 4.44EE675 pKa = 4.74NSAASSSISGYY686 pKa = 8.91TPTFKK691 pKa = 9.95KK692 pKa = 9.22TFEE695 pKa = 4.11ASGQISASATLALPVSLNVGIEE717 pKa = 3.97ITPLSYY723 pKa = 11.0KK724 pKa = 9.36KK725 pKa = 8.74TLSLIEE731 pKa = 4.01QPSLYY736 pKa = 10.69GNVSFAGSAGGADD749 pKa = 3.58AASDD753 pKa = 3.53TCNNGFQYY761 pKa = 10.2FASAQNDD768 pKa = 3.21VSFDD772 pKa = 3.39FFGIKK777 pKa = 9.28TFTLNHH783 pKa = 6.4YY784 pKa = 9.13DD785 pKa = 4.06SPPIQQGCKK794 pKa = 10.34LLDD797 pKa = 4.39DD798 pKa = 4.64SSSTSSSTPAASSGTSSAVTEE819 pKa = 4.68TPSSTGDD826 pKa = 3.18AAADD830 pKa = 3.86SNTATSSGTDD840 pKa = 3.47STSTGAPSPTDD851 pKa = 3.36DD852 pKa = 4.38AAATSTTAGSGTDD865 pKa = 3.25VPTNTGPTSTTDD877 pKa = 3.15TDD879 pKa = 4.09PAPTPSNPTTGDD891 pKa = 3.44STSSKK896 pKa = 10.39SKK898 pKa = 10.66RR899 pKa = 11.84QITDD903 pKa = 2.6SGTVIDD909 pKa = 6.06DD910 pKa = 4.16DD911 pKa = 6.46DD912 pKa = 5.29EE913 pKa = 4.48DD914 pKa = 4.47TFASDD919 pKa = 5.9DD920 pKa = 3.77GDD922 pKa = 4.01EE923 pKa = 4.5TDD925 pKa = 3.95STDD928 pKa = 5.02DD929 pKa = 4.01PVDD932 pKa = 3.96AQDD935 pKa = 5.2SSTDD939 pKa = 3.64PDD941 pKa = 3.51NAEE944 pKa = 4.03YY945 pKa = 9.23TQDD948 pKa = 3.68SSAALNATDD957 pKa = 4.29NAGSDD962 pKa = 3.34GGLTFVTINEE972 pKa = 3.91IHH974 pKa = 6.61NAFQLVSDD982 pKa = 4.29EE983 pKa = 4.42NGNLYY988 pKa = 10.66AGAPSTGDD996 pKa = 3.48DD997 pKa = 4.03DD998 pKa = 4.65GASLFAVADD1007 pKa = 3.96TVVVGNDD1014 pKa = 2.82GDD1016 pKa = 4.51MILHH1020 pKa = 6.58YY1021 pKa = 10.73YY1022 pKa = 9.09PDD1024 pKa = 3.49VMDD1027 pKa = 5.36AYY1029 pKa = 8.58NTSRR1033 pKa = 11.84IRR1035 pKa = 11.84MSYY1038 pKa = 9.89EE1039 pKa = 3.24DD1040 pKa = 5.42HH1041 pKa = 6.54IPKK1044 pKa = 10.43GSEE1047 pKa = 3.87LVTLMALDD1055 pKa = 4.25YY1056 pKa = 11.44DD1057 pKa = 4.56SDD1059 pKa = 4.1DD1060 pKa = 4.3SSNQNILFAVDD1071 pKa = 3.32TKK1073 pKa = 11.59GNAFSLVLCDD1083 pKa = 4.86FSNGADD1089 pKa = 3.03SKK1091 pKa = 11.44LFIVRR1096 pKa = 11.84DD1097 pKa = 3.74TTGVEE1102 pKa = 3.85NLTLEE1107 pKa = 4.21KK1108 pKa = 10.47LRR1110 pKa = 11.84YY1111 pKa = 8.23ICTGAPVSQCDD1122 pKa = 3.4AVALVLGATSTNSS1135 pKa = 2.96
MM1 pKa = 7.55KK2 pKa = 10.21SVSSLSITLLLGLATGSVGLPATTEE27 pKa = 4.56DD28 pKa = 4.17GDD30 pKa = 3.95SHH32 pKa = 6.74GWYY35 pKa = 9.78DD36 pKa = 4.01WNPSTAVTSTSTSFSLSVPSSLPGFAGAPASGFGNTNSNGFAPSVSIKK84 pKa = 9.28TVEE87 pKa = 4.07VTKK90 pKa = 10.86TEE92 pKa = 4.16AVCAATEE99 pKa = 4.01EE100 pKa = 4.79SVVGVGTSTGGSDD113 pKa = 3.47SSHH116 pKa = 6.56SGSGSDD122 pKa = 3.34SSSGSGSGSGSGFVSSSGGSWSEE145 pKa = 4.1SGSTYY150 pKa = 10.31YY151 pKa = 10.95AQAPGANSSLSSPTYY166 pKa = 8.86QPYY169 pKa = 9.86VAGPTLLPAVHH180 pKa = 7.14WDD182 pKa = 3.62YY183 pKa = 11.35PVDD186 pKa = 4.73DD187 pKa = 4.79IRR189 pKa = 11.84NLGPINSSDD198 pKa = 4.4LYY200 pKa = 9.15YY201 pKa = 10.38TSNGIADD208 pKa = 4.15PGVQHH213 pKa = 6.38VFASLSTTLQYY224 pKa = 11.31DD225 pKa = 3.91AVVLDD230 pKa = 3.68HH231 pKa = 7.07SSYY234 pKa = 8.97VTSVSCSSTGILVAFEE250 pKa = 4.39SIEE253 pKa = 3.99AFRR256 pKa = 11.84YY257 pKa = 10.36ACDD260 pKa = 2.75SWSAVEE266 pKa = 4.4EE267 pKa = 4.44FVLVTYY273 pKa = 10.52TDD275 pKa = 3.43GCGASDD281 pKa = 4.01DD282 pKa = 4.64QRR284 pKa = 11.84TFWLSNSVEE293 pKa = 4.21ILNNTQSIMVAVEE306 pKa = 3.95TEE308 pKa = 4.05LAIEE312 pKa = 4.01DD313 pKa = 4.59AIYY316 pKa = 10.23GVDD319 pKa = 3.83MVWGNYY325 pKa = 8.96YY326 pKa = 8.44PTGSNSSTSISGSGSGSSSNSTAPSGSGSNSTGLSGSGTNSTVGVNGTTSSNSTSNANGGSSCGAAPSSTIDD398 pKa = 3.41GFPAVACGVSDD409 pKa = 5.89FDD411 pKa = 3.99TQLDD415 pKa = 4.05DD416 pKa = 3.48VLGYY420 pKa = 11.0LDD422 pKa = 5.8FSSTDD427 pKa = 3.23YY428 pKa = 11.22DD429 pKa = 3.81ASLEE433 pKa = 4.09DD434 pKa = 4.27FSPDD438 pKa = 3.03VSFDD442 pKa = 4.5AEE444 pKa = 5.08DD445 pKa = 5.4LDD447 pKa = 6.5DD448 pKa = 6.62DD449 pKa = 4.61DD450 pKa = 4.7ATMEE454 pKa = 3.98RR455 pKa = 11.84RR456 pKa = 11.84ALLGRR461 pKa = 11.84SHH463 pKa = 6.53LRR465 pKa = 11.84KK466 pKa = 9.87RR467 pKa = 11.84GIFSALLSVGEE478 pKa = 4.1QLSSAVVAVATTGVALARR496 pKa = 11.84AAVQVIPGVSNFIASATEE514 pKa = 3.69FDD516 pKa = 4.2PAVHH520 pKa = 6.04GTATLALAPDD530 pKa = 4.85DD531 pKa = 4.54SATSPWGAAAQLYY544 pKa = 7.79TSSSASATTAADD556 pKa = 2.96ITLYY560 pKa = 10.68CVNCGLKK567 pKa = 10.59GHH569 pKa = 5.7VTLSGSAKK577 pKa = 10.2FNVIDD582 pKa = 4.36GLHH585 pKa = 6.18GLSTSINADD594 pKa = 3.36LEE596 pKa = 4.61AGLNLGLVAQATFKK610 pKa = 10.04DD611 pKa = 4.03TKK613 pKa = 9.51TKK615 pKa = 11.0SLIKK619 pKa = 10.26QAIPDD624 pKa = 3.8LGVSVKK630 pKa = 10.5GVFAAGVYY638 pKa = 9.81IAVDD642 pKa = 3.78AVATLDD648 pKa = 4.89IEE650 pKa = 4.56AQGQALIGITMTISDD665 pKa = 4.37FQATLNLLDD674 pKa = 4.44EE675 pKa = 4.74NSAASSSISGYY686 pKa = 8.91TPTFKK691 pKa = 9.95KK692 pKa = 9.22TFEE695 pKa = 4.11ASGQISASATLALPVSLNVGIEE717 pKa = 3.97ITPLSYY723 pKa = 11.0KK724 pKa = 9.36KK725 pKa = 8.74TLSLIEE731 pKa = 4.01QPSLYY736 pKa = 10.69GNVSFAGSAGGADD749 pKa = 3.58AASDD753 pKa = 3.53TCNNGFQYY761 pKa = 10.2FASAQNDD768 pKa = 3.21VSFDD772 pKa = 3.39FFGIKK777 pKa = 9.28TFTLNHH783 pKa = 6.4YY784 pKa = 9.13DD785 pKa = 4.06SPPIQQGCKK794 pKa = 10.34LLDD797 pKa = 4.39DD798 pKa = 4.64SSSTSSSTPAASSGTSSAVTEE819 pKa = 4.68TPSSTGDD826 pKa = 3.18AAADD830 pKa = 3.86SNTATSSGTDD840 pKa = 3.47STSTGAPSPTDD851 pKa = 3.36DD852 pKa = 4.38AAATSTTAGSGTDD865 pKa = 3.25VPTNTGPTSTTDD877 pKa = 3.15TDD879 pKa = 4.09PAPTPSNPTTGDD891 pKa = 3.44STSSKK896 pKa = 10.39SKK898 pKa = 10.66RR899 pKa = 11.84QITDD903 pKa = 2.6SGTVIDD909 pKa = 6.06DD910 pKa = 4.16DD911 pKa = 6.46DD912 pKa = 5.29EE913 pKa = 4.48DD914 pKa = 4.47TFASDD919 pKa = 5.9DD920 pKa = 3.77GDD922 pKa = 4.01EE923 pKa = 4.5TDD925 pKa = 3.95STDD928 pKa = 5.02DD929 pKa = 4.01PVDD932 pKa = 3.96AQDD935 pKa = 5.2SSTDD939 pKa = 3.64PDD941 pKa = 3.51NAEE944 pKa = 4.03YY945 pKa = 9.23TQDD948 pKa = 3.68SSAALNATDD957 pKa = 4.29NAGSDD962 pKa = 3.34GGLTFVTINEE972 pKa = 3.91IHH974 pKa = 6.61NAFQLVSDD982 pKa = 4.29EE983 pKa = 4.42NGNLYY988 pKa = 10.66AGAPSTGDD996 pKa = 3.48DD997 pKa = 4.03DD998 pKa = 4.65GASLFAVADD1007 pKa = 3.96TVVVGNDD1014 pKa = 2.82GDD1016 pKa = 4.51MILHH1020 pKa = 6.58YY1021 pKa = 10.73YY1022 pKa = 9.09PDD1024 pKa = 3.49VMDD1027 pKa = 5.36AYY1029 pKa = 8.58NTSRR1033 pKa = 11.84IRR1035 pKa = 11.84MSYY1038 pKa = 9.89EE1039 pKa = 3.24DD1040 pKa = 5.42HH1041 pKa = 6.54IPKK1044 pKa = 10.43GSEE1047 pKa = 3.87LVTLMALDD1055 pKa = 4.25YY1056 pKa = 11.44DD1057 pKa = 4.56SDD1059 pKa = 4.1DD1060 pKa = 4.3SSNQNILFAVDD1071 pKa = 3.32TKK1073 pKa = 11.59GNAFSLVLCDD1083 pKa = 4.86FSNGADD1089 pKa = 3.03SKK1091 pKa = 11.44LFIVRR1096 pKa = 11.84DD1097 pKa = 3.74TTGVEE1102 pKa = 3.85NLTLEE1107 pKa = 4.21KK1108 pKa = 10.47LRR1110 pKa = 11.84YY1111 pKa = 8.23ICTGAPVSQCDD1122 pKa = 3.4AVALVLGATSTNSS1135 pKa = 2.96
Molecular weight: 115.62 kDa
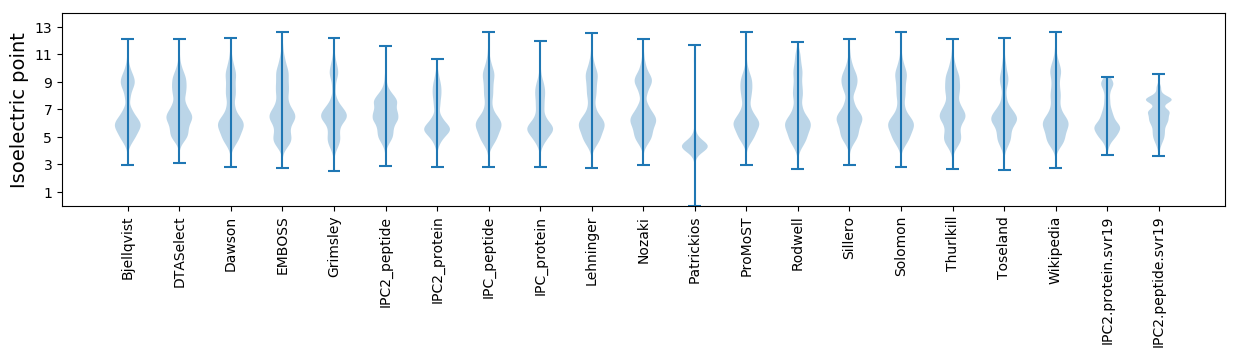
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D1YVL4|A0A0D1YVL4_9EURO Isoform of A0A0D1XEV3 Uncharacterized protein OS=Exophiala sideris OX=1016849 GN=PV11_02182 PE=4 SV=1
MM1 pKa = 7.67SILGSRR7 pKa = 11.84SGILWLRR14 pKa = 11.84SARR17 pKa = 11.84PFVLSICAVFSSSLSEE33 pKa = 3.82HH34 pKa = 6.03LRR36 pKa = 11.84YY37 pKa = 9.6HH38 pKa = 6.46LSCRR42 pKa = 11.84GTTFWQQGRR51 pKa = 11.84VRR53 pKa = 11.84LPPRR57 pKa = 11.84TYY59 pKa = 10.42KK60 pKa = 10.43ISQTYY65 pKa = 10.17ILTSSTGCLKK75 pKa = 10.67PSTILSFGLPFSTKK89 pKa = 9.02PWGCLRR95 pKa = 11.84RR96 pKa = 11.84CRR98 pKa = 11.84HH99 pKa = 5.46NVPIPSLRR107 pKa = 11.84WTRR110 pKa = 11.84KK111 pKa = 8.9VCSLNHH117 pKa = 6.21KK118 pKa = 8.26EE119 pKa = 4.14KK120 pKa = 10.59KK121 pKa = 9.92EE122 pKa = 3.87KK123 pKa = 10.37FSEE126 pKa = 3.97VGEE129 pKa = 4.47NYY131 pKa = 9.61GQPAHH136 pKa = 6.42VPEE139 pKa = 4.35SSPRR143 pKa = 11.84DD144 pKa = 3.44RR145 pKa = 11.84HH146 pKa = 5.04QVSSLDD152 pKa = 3.47VTMGASISRR161 pKa = 11.84PSVHH165 pKa = 6.3SLVVGAHH172 pKa = 6.22SADD175 pKa = 3.07GMYY178 pKa = 10.27QIIDD182 pKa = 3.67GCRR185 pKa = 11.84PNASVYY191 pKa = 9.84PPPGHH196 pKa = 6.83RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84ASRR202 pKa = 11.84TSRR205 pKa = 11.84PSSEE209 pKa = 4.27SASLLL214 pKa = 3.74
MM1 pKa = 7.67SILGSRR7 pKa = 11.84SGILWLRR14 pKa = 11.84SARR17 pKa = 11.84PFVLSICAVFSSSLSEE33 pKa = 3.82HH34 pKa = 6.03LRR36 pKa = 11.84YY37 pKa = 9.6HH38 pKa = 6.46LSCRR42 pKa = 11.84GTTFWQQGRR51 pKa = 11.84VRR53 pKa = 11.84LPPRR57 pKa = 11.84TYY59 pKa = 10.42KK60 pKa = 10.43ISQTYY65 pKa = 10.17ILTSSTGCLKK75 pKa = 10.67PSTILSFGLPFSTKK89 pKa = 9.02PWGCLRR95 pKa = 11.84RR96 pKa = 11.84CRR98 pKa = 11.84HH99 pKa = 5.46NVPIPSLRR107 pKa = 11.84WTRR110 pKa = 11.84KK111 pKa = 8.9VCSLNHH117 pKa = 6.21KK118 pKa = 8.26EE119 pKa = 4.14KK120 pKa = 10.59KK121 pKa = 9.92EE122 pKa = 3.87KK123 pKa = 10.37FSEE126 pKa = 3.97VGEE129 pKa = 4.47NYY131 pKa = 9.61GQPAHH136 pKa = 6.42VPEE139 pKa = 4.35SSPRR143 pKa = 11.84DD144 pKa = 3.44RR145 pKa = 11.84HH146 pKa = 5.04QVSSLDD152 pKa = 3.47VTMGASISRR161 pKa = 11.84PSVHH165 pKa = 6.3SLVVGAHH172 pKa = 6.22SADD175 pKa = 3.07GMYY178 pKa = 10.27QIIDD182 pKa = 3.67GCRR185 pKa = 11.84PNASVYY191 pKa = 9.84PPPGHH196 pKa = 6.83RR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84ASRR202 pKa = 11.84TSRR205 pKa = 11.84PSSEE209 pKa = 4.27SASLLL214 pKa = 3.74
Molecular weight: 23.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5408673 |
50 |
5532 |
498.9 |
55.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.584 ± 0.019 | 1.183 ± 0.01 |
5.81 ± 0.017 | 6.121 ± 0.022 |
3.642 ± 0.012 | 6.728 ± 0.021 |
2.447 ± 0.011 | 4.828 ± 0.015 |
4.895 ± 0.021 | 8.804 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.009 | 3.665 ± 0.013 |
5.991 ± 0.021 | 4.235 ± 0.014 |
6.061 ± 0.021 | 8.263 ± 0.026 |
6.229 ± 0.022 | 6.111 ± 0.014 |
1.421 ± 0.008 | 2.755 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
