
Tatumella ptyseos ATCC 33301
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Erwiniaceae; Tatumella; Tatumella ptyseos
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
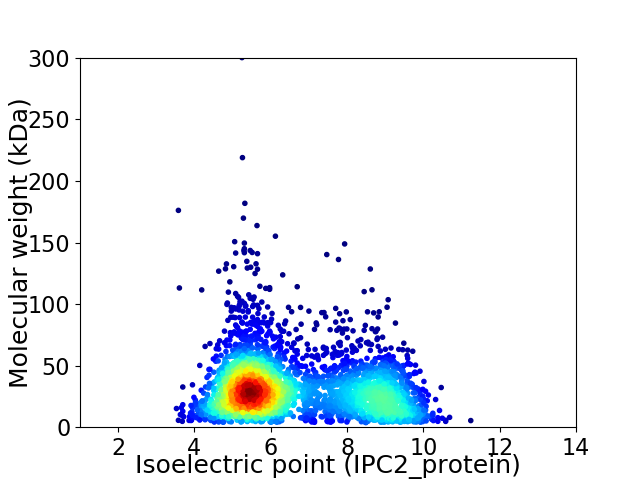
Virtual 2D-PAGE plot for 3337 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A085JL87|A0A085JL87_9GAMM Tyrosine recombinase XerC OS=Tatumella ptyseos ATCC 33301 OX=1005995 GN=xerC PE=3 SV=1
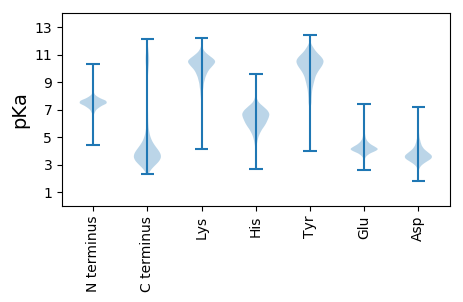
TT1 pKa = 7.4PPVATDD7 pKa = 4.4DD8 pKa = 3.26IAYY11 pKa = 9.02PEE13 pKa = 4.55KK14 pKa = 9.94NTPVSGNVLTNDD26 pKa = 3.35TDD28 pKa = 4.22KK29 pKa = 11.65DD30 pKa = 3.81GDD32 pKa = 4.12PLAVTQFTVEE42 pKa = 3.98NQTAKK47 pKa = 10.83AGATLTLSGVGTLTISSDD65 pKa = 3.06GAYY68 pKa = 9.39TFTPVTGYY76 pKa = 8.91TGPVPSARR84 pKa = 11.84YY85 pKa = 7.91TITDD89 pKa = 3.65GQATATAVLHH99 pKa = 5.75FTDD102 pKa = 3.87ITTGSNTSPVAEE114 pKa = 5.33DD115 pKa = 3.23DD116 pKa = 4.07HH117 pKa = 8.67AVTKK121 pKa = 10.88ADD123 pKa = 3.52TAVSGNVLDD132 pKa = 4.7NDD134 pKa = 3.75SDD136 pKa = 4.42KK137 pKa = 11.72DD138 pKa = 3.8GDD140 pKa = 4.08TLTVTGFSVAGQTLKK155 pKa = 11.02AGEE158 pKa = 4.17TATLDD163 pKa = 3.6GKK165 pKa = 10.05GTLTIGSNGAYY176 pKa = 9.0TFTPLTGYY184 pKa = 8.11TGPVPSATYY193 pKa = 9.67TITDD197 pKa = 3.74GQATSSATLSFSAINTPPVAIADD220 pKa = 3.81HH221 pKa = 6.39AVIDD225 pKa = 3.96GDD227 pKa = 4.22TPVSGNVLSNDD238 pKa = 3.18TDD240 pKa = 4.03KK241 pKa = 11.75DD242 pKa = 3.69GDD244 pKa = 4.2TLTVTQFTVDD254 pKa = 3.5GQTVKK259 pKa = 10.82AGEE262 pKa = 4.04TATISGVGTLTISSDD277 pKa = 2.9GTYY280 pKa = 10.32TFTPVAGYY288 pKa = 8.51TEE290 pKa = 4.45TVPTVSYY297 pKa = 10.21TITDD301 pKa = 3.92GQATASATLIFSDD314 pKa = 3.48TRR316 pKa = 11.84TPPVMAVDD324 pKa = 5.79DD325 pKa = 3.94IAHH328 pKa = 6.92PEE330 pKa = 4.1KK331 pKa = 10.76NSPVTGNVLSNDD343 pKa = 3.15TDD345 pKa = 3.96KK346 pKa = 11.79NGGSLTVTRR355 pKa = 11.84FTVNDD360 pKa = 3.34QTLTAGEE367 pKa = 4.45SATIHH372 pKa = 5.51GVGVLTINANGDD384 pKa = 3.57YY385 pKa = 10.1TFSPEE390 pKa = 3.39IGYY393 pKa = 10.16AGPVPTATYY402 pKa = 9.4TVTNGQSTTSATLSFTDD419 pKa = 5.49IIDD422 pKa = 4.18NPPDD426 pKa = 3.68AVDD429 pKa = 3.58DD430 pKa = 4.04HH431 pKa = 8.03AIVQKK436 pKa = 9.79NTTLSGNALGNDD448 pKa = 3.36SDD450 pKa = 4.76KK451 pKa = 11.72DD452 pKa = 3.58GDD454 pKa = 4.07ALTITDD460 pKa = 4.05FTVNHH465 pKa = 4.99QTFKK469 pKa = 10.94AGEE472 pKa = 3.97TATISGVGTLTFSTSGQYY490 pKa = 10.4IFTPVKK496 pKa = 10.29GYY498 pKa = 10.4VGPVPAATYY507 pKa = 8.27TVSDD511 pKa = 3.98GQLTSQATVYY521 pKa = 10.68FSDD524 pKa = 4.19IKK526 pKa = 10.23NTAPTAVDD534 pKa = 3.29DD535 pKa = 4.53HH536 pKa = 6.41ATTVEE541 pKa = 4.0NTPVSGNVLSNDD553 pKa = 3.25SDD555 pKa = 3.72QDD557 pKa = 3.67GDD559 pKa = 4.25SLTVTQFTVAGQTLSAGKK577 pKa = 7.93TATLSGVGTLTIGSDD592 pKa = 3.09GGYY595 pKa = 8.22TFTPATGYY603 pKa = 8.19TGPVPSVNYY612 pKa = 9.66TITDD616 pKa = 3.79GQATSSATLSFSAINTPPVANNDD639 pKa = 3.51TAVAKK644 pKa = 10.61EE645 pKa = 3.88NTAVSGNVLSNDD657 pKa = 3.18TDD659 pKa = 3.99KK660 pKa = 11.75DD661 pKa = 3.61GDD663 pKa = 4.21TLSVTQFTVDD673 pKa = 3.53GQTLSAGQTATLSGVGTLTIGSNGDD698 pKa = 3.35YY699 pKa = 10.46TFTPVTDD706 pKa = 3.52YY707 pKa = 10.69TGPVPSATYY716 pKa = 9.67TITDD720 pKa = 3.74GQATSSAMLSFSAINTPPVAEE741 pKa = 4.72NDD743 pKa = 3.36TAAPEE748 pKa = 4.35KK749 pKa = 11.05NSTTTGNVLTNDD761 pKa = 3.23SDD763 pKa = 3.99KK764 pKa = 11.71DD765 pKa = 3.56HH766 pKa = 7.42DD767 pKa = 4.32TLSVTTFTVNNQTLNAGEE785 pKa = 4.14TATISGVGTLTIDD798 pKa = 3.58SNGDD802 pKa = 3.29YY803 pKa = 10.87SFTPVTGYY811 pKa = 9.18TGAVPSAKK819 pKa = 9.19YY820 pKa = 9.19TITDD824 pKa = 3.71GQATATATLSFNNIVNTPPSPQDD847 pKa = 3.2DD848 pKa = 4.25TISVQEE854 pKa = 4.08NTPATGNVLSNDD866 pKa = 3.3SDD868 pKa = 4.22KK869 pKa = 11.74DD870 pKa = 3.61GDD872 pKa = 4.21TLTVTQFTIDD882 pKa = 3.64GQTLKK887 pKa = 10.95AGEE890 pKa = 4.22TATLDD895 pKa = 4.22GIGTLTIGSDD905 pKa = 3.57GAFTFTPEE913 pKa = 3.54KK914 pKa = 10.25GYY916 pKa = 9.19TGSVPSATYY925 pKa = 9.44TVTDD929 pKa = 4.04GQTTASAHH937 pKa = 6.68LNIQDD942 pKa = 3.22ITQTYY947 pKa = 8.47TNTPPVANDD956 pKa = 3.2DD957 pKa = 3.86TAVAEE962 pKa = 4.34EE963 pKa = 4.06NTAVSGNVLTNDD975 pKa = 3.29TDD977 pKa = 4.15KK978 pKa = 11.69DD979 pKa = 3.82GDD981 pKa = 4.2TLTVTQFTVDD991 pKa = 3.77GQTLTAGQTATINGVGTLTIGSDD1014 pKa = 3.22GTYY1017 pKa = 10.1TFTPVTGYY1025 pKa = 9.14TGAVPSASYY1034 pKa = 10.03TITDD1038 pKa = 3.86GQATASATLSFSAINTPPVANNDD1061 pKa = 3.21TATAKK1066 pKa = 10.67EE1067 pKa = 3.95NTAVSGNVLSNDD1079 pKa = 3.18TDD1081 pKa = 4.03KK1082 pKa = 11.75DD1083 pKa = 3.69GDD1085 pKa = 4.03TLTVTGFTVDD1095 pKa = 3.64GQTLTAGEE1103 pKa = 4.3TATLSGVGTLTIGSDD1118 pKa = 3.35GAYY1121 pKa = 9.28TFTPVTGYY1129 pKa = 9.14TGAVPSATYY1138 pKa = 9.69TITDD1142 pKa = 3.77GQATASAVLSFTAINTPPVAVNDD1165 pKa = 3.86YY1166 pKa = 9.28VTVDD1170 pKa = 3.17KK1171 pKa = 11.01DD1172 pKa = 3.95SPVSGNVLNNDD1183 pKa = 3.27TDD1185 pKa = 4.18KK1186 pKa = 11.72DD1187 pKa = 3.87GDD1189 pKa = 4.21TLSVTQFSVDD1199 pKa = 3.51GQTLNAGEE1207 pKa = 4.26TATLSGVGTLTIGSDD1222 pKa = 3.35GAYY1225 pKa = 9.37TFTPVSGYY1233 pKa = 10.63VGAVPAATYY1242 pKa = 8.43TISDD1246 pKa = 3.86GQSTAEE1252 pKa = 3.63ASVIFDD1258 pKa = 3.9HH1259 pKa = 7.15APVIEE1264 pKa = 4.34ITQSTEE1270 pKa = 3.42VTYY1273 pKa = 10.41TPGTDD1278 pKa = 2.59ISGFNSSSWQNNWTSAGWKK1297 pKa = 9.43GVWPSYY1303 pKa = 10.75SSTSHH1308 pKa = 6.54LVNLSADD1315 pKa = 3.5GGSYY1319 pKa = 9.51TGTMNAATTFNTNDD1333 pKa = 3.46VLDD1336 pKa = 4.48LTVGWNNGWSGTDD1349 pKa = 3.5DD1350 pKa = 3.67GSSAVLSFSYY1360 pKa = 8.82GTQTLFSITTPGQSTTGTTPGAEE1383 pKa = 4.0YY1384 pKa = 11.18ADD1386 pKa = 3.83AQKK1389 pKa = 10.84YY1390 pKa = 10.38AYY1392 pKa = 7.64ITLSKK1397 pKa = 10.39GIEE1400 pKa = 3.83VSLDD1404 pKa = 3.35GGKK1407 pKa = 7.68TWLDD1411 pKa = 3.24GSTPIPFEE1419 pKa = 3.72TWRR1422 pKa = 11.84LYY1424 pKa = 10.9QKK1426 pKa = 10.64DD1427 pKa = 3.64QTVTDD1432 pKa = 4.0SDD1434 pKa = 4.02LSHH1437 pKa = 7.71ILVKK1441 pKa = 10.69GMQTGVSDD1449 pKa = 4.2HH1450 pKa = 7.05LVLNWDD1456 pKa = 4.13APGSPADD1463 pKa = 4.16DD1464 pKa = 3.8FQISGVSVSQVTEE1477 pKa = 4.13TTSDD1481 pKa = 3.59SDD1483 pKa = 4.02SHH1485 pKa = 6.11TLTYY1489 pKa = 10.76DD1490 pKa = 3.41PSTSSGTDD1498 pKa = 3.17STSSQLAVLPNISVSDD1514 pKa = 3.67SDD1516 pKa = 4.14GDD1518 pKa = 3.97NDD1520 pKa = 3.73IQSATITLSGKK1531 pKa = 10.47SGDD1534 pKa = 3.89QLWINGMDD1542 pKa = 3.77ASTHH1546 pKa = 4.75TAGKK1550 pKa = 10.2ASYY1553 pKa = 10.57SLTSDD1558 pKa = 3.78GEE1560 pKa = 4.51GTLTLTLKK1568 pKa = 10.81AGTSGSLSSSDD1579 pKa = 2.95WNTLLEE1585 pKa = 4.04GLRR1588 pKa = 11.84FGTTGDD1594 pKa = 3.06SGTRR1598 pKa = 11.84QLTVTVNDD1606 pKa = 4.23GQLDD1610 pKa = 4.41SNTEE1614 pKa = 4.03SVDD1617 pKa = 3.31INVTGSASSGDD1628 pKa = 3.78LPGYY1632 pKa = 10.05SADD1635 pKa = 3.79TQTTTTSSLLSTYY1648 pKa = 10.14SLEE1651 pKa = 4.28NVSDD1655 pKa = 3.96LASASDD1661 pKa = 3.58TSSASTTEE1669 pKa = 4.03TLPTLQSLLSTSSTTTSSTGSSASSTTASSSSATDD1704 pKa = 3.44SLHH1707 pKa = 5.21TTSEE1711 pKa = 4.17TSSALLAKK1719 pKa = 10.22EE1720 pKa = 5.21DD1721 pKa = 3.98EE1722 pKa = 4.54LSSNNNTVV1730 pKa = 2.58
TT1 pKa = 7.4PPVATDD7 pKa = 4.4DD8 pKa = 3.26IAYY11 pKa = 9.02PEE13 pKa = 4.55KK14 pKa = 9.94NTPVSGNVLTNDD26 pKa = 3.35TDD28 pKa = 4.22KK29 pKa = 11.65DD30 pKa = 3.81GDD32 pKa = 4.12PLAVTQFTVEE42 pKa = 3.98NQTAKK47 pKa = 10.83AGATLTLSGVGTLTISSDD65 pKa = 3.06GAYY68 pKa = 9.39TFTPVTGYY76 pKa = 8.91TGPVPSARR84 pKa = 11.84YY85 pKa = 7.91TITDD89 pKa = 3.65GQATATAVLHH99 pKa = 5.75FTDD102 pKa = 3.87ITTGSNTSPVAEE114 pKa = 5.33DD115 pKa = 3.23DD116 pKa = 4.07HH117 pKa = 8.67AVTKK121 pKa = 10.88ADD123 pKa = 3.52TAVSGNVLDD132 pKa = 4.7NDD134 pKa = 3.75SDD136 pKa = 4.42KK137 pKa = 11.72DD138 pKa = 3.8GDD140 pKa = 4.08TLTVTGFSVAGQTLKK155 pKa = 11.02AGEE158 pKa = 4.17TATLDD163 pKa = 3.6GKK165 pKa = 10.05GTLTIGSNGAYY176 pKa = 9.0TFTPLTGYY184 pKa = 8.11TGPVPSATYY193 pKa = 9.67TITDD197 pKa = 3.74GQATSSATLSFSAINTPPVAIADD220 pKa = 3.81HH221 pKa = 6.39AVIDD225 pKa = 3.96GDD227 pKa = 4.22TPVSGNVLSNDD238 pKa = 3.18TDD240 pKa = 4.03KK241 pKa = 11.75DD242 pKa = 3.69GDD244 pKa = 4.2TLTVTQFTVDD254 pKa = 3.5GQTVKK259 pKa = 10.82AGEE262 pKa = 4.04TATISGVGTLTISSDD277 pKa = 2.9GTYY280 pKa = 10.32TFTPVAGYY288 pKa = 8.51TEE290 pKa = 4.45TVPTVSYY297 pKa = 10.21TITDD301 pKa = 3.92GQATASATLIFSDD314 pKa = 3.48TRR316 pKa = 11.84TPPVMAVDD324 pKa = 5.79DD325 pKa = 3.94IAHH328 pKa = 6.92PEE330 pKa = 4.1KK331 pKa = 10.76NSPVTGNVLSNDD343 pKa = 3.15TDD345 pKa = 3.96KK346 pKa = 11.79NGGSLTVTRR355 pKa = 11.84FTVNDD360 pKa = 3.34QTLTAGEE367 pKa = 4.45SATIHH372 pKa = 5.51GVGVLTINANGDD384 pKa = 3.57YY385 pKa = 10.1TFSPEE390 pKa = 3.39IGYY393 pKa = 10.16AGPVPTATYY402 pKa = 9.4TVTNGQSTTSATLSFTDD419 pKa = 5.49IIDD422 pKa = 4.18NPPDD426 pKa = 3.68AVDD429 pKa = 3.58DD430 pKa = 4.04HH431 pKa = 8.03AIVQKK436 pKa = 9.79NTTLSGNALGNDD448 pKa = 3.36SDD450 pKa = 4.76KK451 pKa = 11.72DD452 pKa = 3.58GDD454 pKa = 4.07ALTITDD460 pKa = 4.05FTVNHH465 pKa = 4.99QTFKK469 pKa = 10.94AGEE472 pKa = 3.97TATISGVGTLTFSTSGQYY490 pKa = 10.4IFTPVKK496 pKa = 10.29GYY498 pKa = 10.4VGPVPAATYY507 pKa = 8.27TVSDD511 pKa = 3.98GQLTSQATVYY521 pKa = 10.68FSDD524 pKa = 4.19IKK526 pKa = 10.23NTAPTAVDD534 pKa = 3.29DD535 pKa = 4.53HH536 pKa = 6.41ATTVEE541 pKa = 4.0NTPVSGNVLSNDD553 pKa = 3.25SDD555 pKa = 3.72QDD557 pKa = 3.67GDD559 pKa = 4.25SLTVTQFTVAGQTLSAGKK577 pKa = 7.93TATLSGVGTLTIGSDD592 pKa = 3.09GGYY595 pKa = 8.22TFTPATGYY603 pKa = 8.19TGPVPSVNYY612 pKa = 9.66TITDD616 pKa = 3.79GQATSSATLSFSAINTPPVANNDD639 pKa = 3.51TAVAKK644 pKa = 10.61EE645 pKa = 3.88NTAVSGNVLSNDD657 pKa = 3.18TDD659 pKa = 3.99KK660 pKa = 11.75DD661 pKa = 3.61GDD663 pKa = 4.21TLSVTQFTVDD673 pKa = 3.53GQTLSAGQTATLSGVGTLTIGSNGDD698 pKa = 3.35YY699 pKa = 10.46TFTPVTDD706 pKa = 3.52YY707 pKa = 10.69TGPVPSATYY716 pKa = 9.67TITDD720 pKa = 3.74GQATSSAMLSFSAINTPPVAEE741 pKa = 4.72NDD743 pKa = 3.36TAAPEE748 pKa = 4.35KK749 pKa = 11.05NSTTTGNVLTNDD761 pKa = 3.23SDD763 pKa = 3.99KK764 pKa = 11.71DD765 pKa = 3.56HH766 pKa = 7.42DD767 pKa = 4.32TLSVTTFTVNNQTLNAGEE785 pKa = 4.14TATISGVGTLTIDD798 pKa = 3.58SNGDD802 pKa = 3.29YY803 pKa = 10.87SFTPVTGYY811 pKa = 9.18TGAVPSAKK819 pKa = 9.19YY820 pKa = 9.19TITDD824 pKa = 3.71GQATATATLSFNNIVNTPPSPQDD847 pKa = 3.2DD848 pKa = 4.25TISVQEE854 pKa = 4.08NTPATGNVLSNDD866 pKa = 3.3SDD868 pKa = 4.22KK869 pKa = 11.74DD870 pKa = 3.61GDD872 pKa = 4.21TLTVTQFTIDD882 pKa = 3.64GQTLKK887 pKa = 10.95AGEE890 pKa = 4.22TATLDD895 pKa = 4.22GIGTLTIGSDD905 pKa = 3.57GAFTFTPEE913 pKa = 3.54KK914 pKa = 10.25GYY916 pKa = 9.19TGSVPSATYY925 pKa = 9.44TVTDD929 pKa = 4.04GQTTASAHH937 pKa = 6.68LNIQDD942 pKa = 3.22ITQTYY947 pKa = 8.47TNTPPVANDD956 pKa = 3.2DD957 pKa = 3.86TAVAEE962 pKa = 4.34EE963 pKa = 4.06NTAVSGNVLTNDD975 pKa = 3.29TDD977 pKa = 4.15KK978 pKa = 11.69DD979 pKa = 3.82GDD981 pKa = 4.2TLTVTQFTVDD991 pKa = 3.77GQTLTAGQTATINGVGTLTIGSDD1014 pKa = 3.22GTYY1017 pKa = 10.1TFTPVTGYY1025 pKa = 9.14TGAVPSASYY1034 pKa = 10.03TITDD1038 pKa = 3.86GQATASATLSFSAINTPPVANNDD1061 pKa = 3.21TATAKK1066 pKa = 10.67EE1067 pKa = 3.95NTAVSGNVLSNDD1079 pKa = 3.18TDD1081 pKa = 4.03KK1082 pKa = 11.75DD1083 pKa = 3.69GDD1085 pKa = 4.03TLTVTGFTVDD1095 pKa = 3.64GQTLTAGEE1103 pKa = 4.3TATLSGVGTLTIGSDD1118 pKa = 3.35GAYY1121 pKa = 9.28TFTPVTGYY1129 pKa = 9.14TGAVPSATYY1138 pKa = 9.69TITDD1142 pKa = 3.77GQATASAVLSFTAINTPPVAVNDD1165 pKa = 3.86YY1166 pKa = 9.28VTVDD1170 pKa = 3.17KK1171 pKa = 11.01DD1172 pKa = 3.95SPVSGNVLNNDD1183 pKa = 3.27TDD1185 pKa = 4.18KK1186 pKa = 11.72DD1187 pKa = 3.87GDD1189 pKa = 4.21TLSVTQFSVDD1199 pKa = 3.51GQTLNAGEE1207 pKa = 4.26TATLSGVGTLTIGSDD1222 pKa = 3.35GAYY1225 pKa = 9.37TFTPVSGYY1233 pKa = 10.63VGAVPAATYY1242 pKa = 8.43TISDD1246 pKa = 3.86GQSTAEE1252 pKa = 3.63ASVIFDD1258 pKa = 3.9HH1259 pKa = 7.15APVIEE1264 pKa = 4.34ITQSTEE1270 pKa = 3.42VTYY1273 pKa = 10.41TPGTDD1278 pKa = 2.59ISGFNSSSWQNNWTSAGWKK1297 pKa = 9.43GVWPSYY1303 pKa = 10.75SSTSHH1308 pKa = 6.54LVNLSADD1315 pKa = 3.5GGSYY1319 pKa = 9.51TGTMNAATTFNTNDD1333 pKa = 3.46VLDD1336 pKa = 4.48LTVGWNNGWSGTDD1349 pKa = 3.5DD1350 pKa = 3.67GSSAVLSFSYY1360 pKa = 8.82GTQTLFSITTPGQSTTGTTPGAEE1383 pKa = 4.0YY1384 pKa = 11.18ADD1386 pKa = 3.83AQKK1389 pKa = 10.84YY1390 pKa = 10.38AYY1392 pKa = 7.64ITLSKK1397 pKa = 10.39GIEE1400 pKa = 3.83VSLDD1404 pKa = 3.35GGKK1407 pKa = 7.68TWLDD1411 pKa = 3.24GSTPIPFEE1419 pKa = 3.72TWRR1422 pKa = 11.84LYY1424 pKa = 10.9QKK1426 pKa = 10.64DD1427 pKa = 3.64QTVTDD1432 pKa = 4.0SDD1434 pKa = 4.02LSHH1437 pKa = 7.71ILVKK1441 pKa = 10.69GMQTGVSDD1449 pKa = 4.2HH1450 pKa = 7.05LVLNWDD1456 pKa = 4.13APGSPADD1463 pKa = 4.16DD1464 pKa = 3.8FQISGVSVSQVTEE1477 pKa = 4.13TTSDD1481 pKa = 3.59SDD1483 pKa = 4.02SHH1485 pKa = 6.11TLTYY1489 pKa = 10.76DD1490 pKa = 3.41PSTSSGTDD1498 pKa = 3.17STSSQLAVLPNISVSDD1514 pKa = 3.67SDD1516 pKa = 4.14GDD1518 pKa = 3.97NDD1520 pKa = 3.73IQSATITLSGKK1531 pKa = 10.47SGDD1534 pKa = 3.89QLWINGMDD1542 pKa = 3.77ASTHH1546 pKa = 4.75TAGKK1550 pKa = 10.2ASYY1553 pKa = 10.57SLTSDD1558 pKa = 3.78GEE1560 pKa = 4.51GTLTLTLKK1568 pKa = 10.81AGTSGSLSSSDD1579 pKa = 2.95WNTLLEE1585 pKa = 4.04GLRR1588 pKa = 11.84FGTTGDD1594 pKa = 3.06SGTRR1598 pKa = 11.84QLTVTVNDD1606 pKa = 4.23GQLDD1610 pKa = 4.41SNTEE1614 pKa = 4.03SVDD1617 pKa = 3.31INVTGSASSGDD1628 pKa = 3.78LPGYY1632 pKa = 10.05SADD1635 pKa = 3.79TQTTTTSSLLSTYY1648 pKa = 10.14SLEE1651 pKa = 4.28NVSDD1655 pKa = 3.96LASASDD1661 pKa = 3.58TSSASTTEE1669 pKa = 4.03TLPTLQSLLSTSSTTTSSTGSSASSTTASSSSATDD1704 pKa = 3.44SLHH1707 pKa = 5.21TTSEE1711 pKa = 4.17TSSALLAKK1719 pKa = 10.22EE1720 pKa = 5.21DD1721 pKa = 3.98EE1722 pKa = 4.54LSSNNNTVV1730 pKa = 2.58
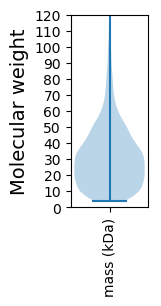
Molecular weight: 176.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085JPE5|A0A085JPE5_9GAMM Uncharacterized protein OS=Tatumella ptyseos ATCC 33301 OX=1005995 GN=GTPT_0515 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1003587 |
37 |
2922 |
300.7 |
33.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.432 ± 0.049 | 1.115 ± 0.016 |
5.12 ± 0.035 | 5.478 ± 0.046 |
3.816 ± 0.03 | 7.541 ± 0.039 |
2.314 ± 0.02 | 5.838 ± 0.038 |
3.938 ± 0.031 | 10.932 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.624 ± 0.023 | 3.54 ± 0.032 |
4.646 ± 0.031 | 4.732 ± 0.043 |
5.888 ± 0.04 | 6.477 ± 0.04 |
5.564 ± 0.038 | 6.758 ± 0.032 |
1.411 ± 0.017 | 2.836 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
