
Equine encephalosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
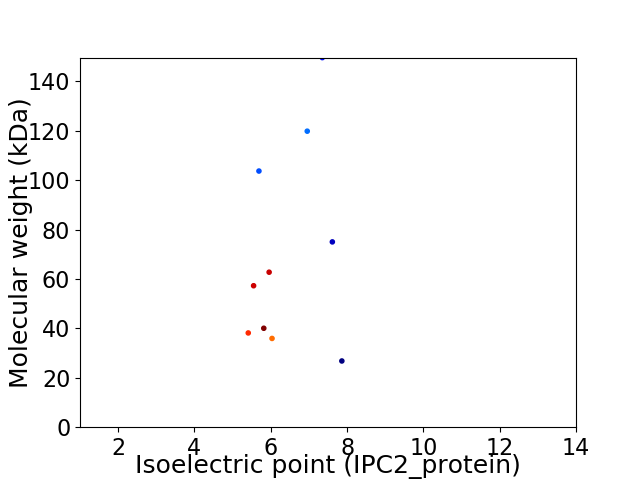
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S6CFI5|S6CFI5_9REOV VP2 protein OS=Equine encephalosis virus OX=201490 GN=VP2 PE=4 SV=1
MM1 pKa = 7.73DD2 pKa = 6.41AIAARR7 pKa = 11.84ALSVVRR13 pKa = 11.84ACVTAQDD20 pKa = 4.38PRR22 pKa = 11.84ATMAPQVLEE31 pKa = 3.84ILGLGINRR39 pKa = 11.84YY40 pKa = 8.24NAITGSRR47 pKa = 11.84VTMRR51 pKa = 11.84PGTVPEE57 pKa = 4.3RR58 pKa = 11.84NDD60 pKa = 3.29MMFMCIDD67 pKa = 4.0MVCAALNIQLGNVSPDD83 pKa = 3.26YY84 pKa = 9.14TQKK87 pKa = 11.27LEE89 pKa = 4.45TIGVLATSEE98 pKa = 3.76IPYY101 pKa = 9.69TDD103 pKa = 3.43EE104 pKa = 5.07AINAIVRR111 pKa = 11.84VTGEE115 pKa = 4.03TQTWGPVDD123 pKa = 4.11SPMPPYY129 pKa = 10.39LGAVAVEE136 pKa = 4.3APGHH140 pKa = 5.29YY141 pKa = 10.07FMPLGRR147 pKa = 11.84QTHH150 pKa = 5.12SAYY153 pKa = 9.56TDD155 pKa = 3.43SNTIQVSLTPNSAAQVNGAMTPQGVEE181 pKa = 3.81LVQLFFVWRR190 pKa = 11.84PFAMYY195 pKa = 9.57TSQAGAAVAQPPGVTLSLGGVNMPEE220 pKa = 3.64GRR222 pKa = 11.84ILIWDD227 pKa = 4.16GVAPVVVQNPGNAPAMAQIEE247 pKa = 4.73VVRR250 pKa = 11.84FTQLLHH256 pKa = 7.54SYY258 pKa = 7.59EE259 pKa = 4.46TVPEE263 pKa = 4.09TRR265 pKa = 11.84EE266 pKa = 3.76NMARR270 pKa = 11.84CYY272 pKa = 11.01SFLSVTWHH280 pKa = 5.58TLRR283 pKa = 11.84SAILRR288 pKa = 11.84SLGMLPVHH296 pKa = 6.37QSAYY300 pKa = 9.69PPTNRR305 pKa = 11.84YY306 pKa = 8.91EE307 pKa = 4.0YY308 pKa = 10.77LAYY311 pKa = 10.96LLIAALADD319 pKa = 3.88AYY321 pKa = 10.83DD322 pKa = 3.81ALRR325 pKa = 11.84PNFDD329 pKa = 4.04MFNQPNVPLQPTRR342 pKa = 11.84AQIAALYY349 pKa = 9.69RR350 pKa = 3.85
MM1 pKa = 7.73DD2 pKa = 6.41AIAARR7 pKa = 11.84ALSVVRR13 pKa = 11.84ACVTAQDD20 pKa = 4.38PRR22 pKa = 11.84ATMAPQVLEE31 pKa = 3.84ILGLGINRR39 pKa = 11.84YY40 pKa = 8.24NAITGSRR47 pKa = 11.84VTMRR51 pKa = 11.84PGTVPEE57 pKa = 4.3RR58 pKa = 11.84NDD60 pKa = 3.29MMFMCIDD67 pKa = 4.0MVCAALNIQLGNVSPDD83 pKa = 3.26YY84 pKa = 9.14TQKK87 pKa = 11.27LEE89 pKa = 4.45TIGVLATSEE98 pKa = 3.76IPYY101 pKa = 9.69TDD103 pKa = 3.43EE104 pKa = 5.07AINAIVRR111 pKa = 11.84VTGEE115 pKa = 4.03TQTWGPVDD123 pKa = 4.11SPMPPYY129 pKa = 10.39LGAVAVEE136 pKa = 4.3APGHH140 pKa = 5.29YY141 pKa = 10.07FMPLGRR147 pKa = 11.84QTHH150 pKa = 5.12SAYY153 pKa = 9.56TDD155 pKa = 3.43SNTIQVSLTPNSAAQVNGAMTPQGVEE181 pKa = 3.81LVQLFFVWRR190 pKa = 11.84PFAMYY195 pKa = 9.57TSQAGAAVAQPPGVTLSLGGVNMPEE220 pKa = 3.64GRR222 pKa = 11.84ILIWDD227 pKa = 4.16GVAPVVVQNPGNAPAMAQIEE247 pKa = 4.73VVRR250 pKa = 11.84FTQLLHH256 pKa = 7.54SYY258 pKa = 7.59EE259 pKa = 4.46TVPEE263 pKa = 4.09TRR265 pKa = 11.84EE266 pKa = 3.76NMARR270 pKa = 11.84CYY272 pKa = 11.01SFLSVTWHH280 pKa = 5.58TLRR283 pKa = 11.84SAILRR288 pKa = 11.84SLGMLPVHH296 pKa = 6.37QSAYY300 pKa = 9.69PPTNRR305 pKa = 11.84YY306 pKa = 8.91EE307 pKa = 4.0YY308 pKa = 10.77LAYY311 pKa = 10.96LLIAALADD319 pKa = 3.88AYY321 pKa = 10.83DD322 pKa = 3.81ALRR325 pKa = 11.84PNFDD329 pKa = 4.04MFNQPNVPLQPTRR342 pKa = 11.84AQIAALYY349 pKa = 9.69RR350 pKa = 3.85
Molecular weight: 38.15 kDa
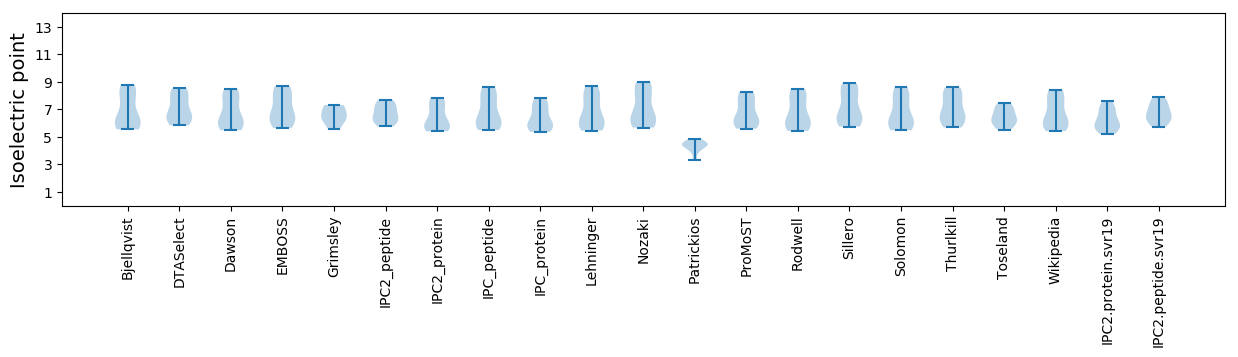
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S6CGJ8|S6CGJ8_9REOV Outer capsid protein VP5 OS=Equine encephalosis virus OX=201490 GN=VP5 PE=3 SV=1
MM1 pKa = 7.13YY2 pKa = 9.04PVLSRR7 pKa = 11.84AVVGNPEE14 pKa = 3.6EE15 pKa = 4.24RR16 pKa = 11.84ALMVYY21 pKa = 10.09PPTAPMPPVTTWDD34 pKa = 3.43NLKK37 pKa = 9.91IDD39 pKa = 4.4SVDD42 pKa = 3.35GMKK45 pKa = 10.74DD46 pKa = 3.01LALNILDD53 pKa = 4.36KK54 pKa = 11.23NITSTTGADD63 pKa = 3.23EE64 pKa = 4.07CDD66 pKa = 3.51KK67 pKa = 11.19RR68 pKa = 11.84EE69 pKa = 3.95KK70 pKa = 11.46AMFASVAEE78 pKa = 4.39SAADD82 pKa = 3.41SPMVRR87 pKa = 11.84TIKK90 pKa = 9.83IQIYY94 pKa = 10.44NRR96 pKa = 11.84VLDD99 pKa = 3.92DD100 pKa = 3.46MEE102 pKa = 4.8RR103 pKa = 11.84EE104 pKa = 3.8KK105 pKa = 10.98RR106 pKa = 11.84KK107 pKa = 9.64CEE109 pKa = 3.58KK110 pKa = 9.93RR111 pKa = 11.84RR112 pKa = 11.84AMLRR116 pKa = 11.84FVSNAFITLMLMSTFLMAMMQTPPITQYY144 pKa = 10.69VEE146 pKa = 4.19KK147 pKa = 10.28ACNGTGGTEE156 pKa = 4.17TNDD159 pKa = 3.3PCGLMRR165 pKa = 11.84WSGAVQFLMLIMSGFLYY182 pKa = 9.47MCKK185 pKa = 9.67RR186 pKa = 11.84WITTLSTNADD196 pKa = 4.29RR197 pKa = 11.84ISKK200 pKa = 10.51NILKK204 pKa = 10.14RR205 pKa = 11.84RR206 pKa = 11.84AYY208 pKa = 9.69IDD210 pKa = 3.39AARR213 pKa = 11.84SNPNATVLTVTGGNTGDD230 pKa = 4.18LPYY233 pKa = 10.86QFGDD237 pKa = 3.51TAHH240 pKa = 6.87
MM1 pKa = 7.13YY2 pKa = 9.04PVLSRR7 pKa = 11.84AVVGNPEE14 pKa = 3.6EE15 pKa = 4.24RR16 pKa = 11.84ALMVYY21 pKa = 10.09PPTAPMPPVTTWDD34 pKa = 3.43NLKK37 pKa = 9.91IDD39 pKa = 4.4SVDD42 pKa = 3.35GMKK45 pKa = 10.74DD46 pKa = 3.01LALNILDD53 pKa = 4.36KK54 pKa = 11.23NITSTTGADD63 pKa = 3.23EE64 pKa = 4.07CDD66 pKa = 3.51KK67 pKa = 11.19RR68 pKa = 11.84EE69 pKa = 3.95KK70 pKa = 11.46AMFASVAEE78 pKa = 4.39SAADD82 pKa = 3.41SPMVRR87 pKa = 11.84TIKK90 pKa = 9.83IQIYY94 pKa = 10.44NRR96 pKa = 11.84VLDD99 pKa = 3.92DD100 pKa = 3.46MEE102 pKa = 4.8RR103 pKa = 11.84EE104 pKa = 3.8KK105 pKa = 10.98RR106 pKa = 11.84KK107 pKa = 9.64CEE109 pKa = 3.58KK110 pKa = 9.93RR111 pKa = 11.84RR112 pKa = 11.84AMLRR116 pKa = 11.84FVSNAFITLMLMSTFLMAMMQTPPITQYY144 pKa = 10.69VEE146 pKa = 4.19KK147 pKa = 10.28ACNGTGGTEE156 pKa = 4.17TNDD159 pKa = 3.3PCGLMRR165 pKa = 11.84WSGAVQFLMLIMSGFLYY182 pKa = 9.47MCKK185 pKa = 9.67RR186 pKa = 11.84WITTLSTNADD196 pKa = 4.29RR197 pKa = 11.84ISKK200 pKa = 10.51NILKK204 pKa = 10.14RR205 pKa = 11.84RR206 pKa = 11.84AYY208 pKa = 9.69IDD210 pKa = 3.39AARR213 pKa = 11.84SNPNATVLTVTGGNTGDD230 pKa = 4.18LPYY233 pKa = 10.86QFGDD237 pKa = 3.51TAHH240 pKa = 6.87
Molecular weight: 26.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6257 |
240 |
1307 |
625.7 |
70.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.249 ± 0.633 | 1.406 ± 0.252 |
5.849 ± 0.21 | 6.856 ± 0.404 |
3.964 ± 0.333 | 6.601 ± 0.666 |
2.317 ± 0.287 | 6.505 ± 0.331 |
5.258 ± 0.604 | 8.471 ± 0.243 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.516 ± 0.209 | 4.235 ± 0.137 |
4.475 ± 0.376 | 3.5 ± 0.284 |
7.032 ± 0.215 | 6.585 ± 0.515 |
5.514 ± 0.317 | 7.08 ± 0.271 |
1.167 ± 0.242 | 3.42 ± 0.231 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
