
Novibacillus thermophilus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Thermoactinomycetaceae; Novibacillus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
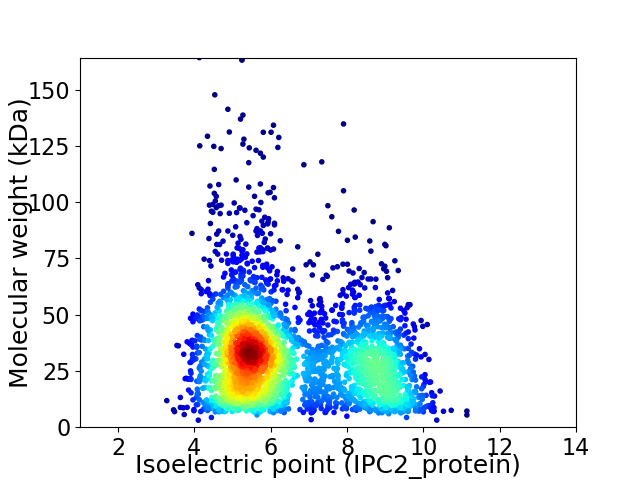
Virtual 2D-PAGE plot for 3169 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1U9K710|A0A1U9K710_9BACL Methionine import ATP-binding protein MetN OS=Novibacillus thermophilus OX=1471761 GN=metN PE=3 SV=1
MM1 pKa = 7.26EE2 pKa = 5.5KK3 pKa = 10.32KK4 pKa = 10.84GEE6 pKa = 4.07FYY8 pKa = 10.91LKK10 pKa = 10.08YY11 pKa = 10.96VKK13 pKa = 9.15ITGLLFIIFALALVGCGSSQDD34 pKa = 4.31DD35 pKa = 3.69SGADD39 pKa = 3.39SNEE42 pKa = 3.88GAEE45 pKa = 4.22EE46 pKa = 3.89TDD48 pKa = 3.57AGSEE52 pKa = 4.14DD53 pKa = 4.12VVEE56 pKa = 4.97GDD58 pKa = 3.68TPDD61 pKa = 3.79FLSMLTGGTSGTYY74 pKa = 9.52YY75 pKa = 10.48PLGGEE80 pKa = 4.04MASIITQEE88 pKa = 4.04TGIQTDD94 pKa = 3.66AVSSNASADD103 pKa = 3.58NIMAVHH109 pKa = 6.29NGEE112 pKa = 4.39AEE114 pKa = 4.21LAFTQTDD121 pKa = 3.36VMANAAEE128 pKa = 4.27GVNAFDD134 pKa = 5.07EE135 pKa = 5.2PIEE138 pKa = 4.35DD139 pKa = 3.59VLAIGSLYY147 pKa = 10.17PEE149 pKa = 5.24TIQIVTTADD158 pKa = 3.0SGINSVEE165 pKa = 4.0DD166 pKa = 3.58LAGKK170 pKa = 7.19TVSVGAPGSGTYY182 pKa = 10.16INAEE186 pKa = 3.98QILEE190 pKa = 4.15VHH192 pKa = 6.3GMTMDD197 pKa = 6.51DD198 pKa = 4.57IDD200 pKa = 4.42AQNLDD205 pKa = 3.86FGEE208 pKa = 4.51STGGIQDD215 pKa = 3.77GNIDD219 pKa = 3.72AAFITAGTPTGAVEE233 pKa = 4.36GLTATTDD240 pKa = 3.11ISIVPVAADD249 pKa = 3.23KK250 pKa = 11.59AEE252 pKa = 3.95ALIEE256 pKa = 4.26KK257 pKa = 9.31YY258 pKa = 9.84PYY260 pKa = 9.8YY261 pKa = 10.49ATDD264 pKa = 3.3VVSAGTYY271 pKa = 9.97GLEE274 pKa = 3.95EE275 pKa = 4.48DD276 pKa = 4.3VEE278 pKa = 4.96TVAVLAMLAVTDD290 pKa = 4.37SLSEE294 pKa = 4.03DD295 pKa = 3.38LVYY298 pKa = 10.85DD299 pKa = 3.37ITKK302 pKa = 10.51AIYY305 pKa = 10.16EE306 pKa = 4.13NTEE309 pKa = 3.87NMSHH313 pKa = 6.53AKK315 pKa = 10.57AEE317 pKa = 4.62FIQVEE322 pKa = 4.27TALDD326 pKa = 4.51GIGIDD331 pKa = 3.64LHH333 pKa = 6.76PGAEE337 pKa = 4.26KK338 pKa = 10.85YY339 pKa = 8.47FTEE342 pKa = 5.04AGVLEE347 pKa = 4.37
MM1 pKa = 7.26EE2 pKa = 5.5KK3 pKa = 10.32KK4 pKa = 10.84GEE6 pKa = 4.07FYY8 pKa = 10.91LKK10 pKa = 10.08YY11 pKa = 10.96VKK13 pKa = 9.15ITGLLFIIFALALVGCGSSQDD34 pKa = 4.31DD35 pKa = 3.69SGADD39 pKa = 3.39SNEE42 pKa = 3.88GAEE45 pKa = 4.22EE46 pKa = 3.89TDD48 pKa = 3.57AGSEE52 pKa = 4.14DD53 pKa = 4.12VVEE56 pKa = 4.97GDD58 pKa = 3.68TPDD61 pKa = 3.79FLSMLTGGTSGTYY74 pKa = 9.52YY75 pKa = 10.48PLGGEE80 pKa = 4.04MASIITQEE88 pKa = 4.04TGIQTDD94 pKa = 3.66AVSSNASADD103 pKa = 3.58NIMAVHH109 pKa = 6.29NGEE112 pKa = 4.39AEE114 pKa = 4.21LAFTQTDD121 pKa = 3.36VMANAAEE128 pKa = 4.27GVNAFDD134 pKa = 5.07EE135 pKa = 5.2PIEE138 pKa = 4.35DD139 pKa = 3.59VLAIGSLYY147 pKa = 10.17PEE149 pKa = 5.24TIQIVTTADD158 pKa = 3.0SGINSVEE165 pKa = 4.0DD166 pKa = 3.58LAGKK170 pKa = 7.19TVSVGAPGSGTYY182 pKa = 10.16INAEE186 pKa = 3.98QILEE190 pKa = 4.15VHH192 pKa = 6.3GMTMDD197 pKa = 6.51DD198 pKa = 4.57IDD200 pKa = 4.42AQNLDD205 pKa = 3.86FGEE208 pKa = 4.51STGGIQDD215 pKa = 3.77GNIDD219 pKa = 3.72AAFITAGTPTGAVEE233 pKa = 4.36GLTATTDD240 pKa = 3.11ISIVPVAADD249 pKa = 3.23KK250 pKa = 11.59AEE252 pKa = 3.95ALIEE256 pKa = 4.26KK257 pKa = 9.31YY258 pKa = 9.84PYY260 pKa = 9.8YY261 pKa = 10.49ATDD264 pKa = 3.3VVSAGTYY271 pKa = 9.97GLEE274 pKa = 3.95EE275 pKa = 4.48DD276 pKa = 4.3VEE278 pKa = 4.96TVAVLAMLAVTDD290 pKa = 4.37SLSEE294 pKa = 4.03DD295 pKa = 3.38LVYY298 pKa = 10.85DD299 pKa = 3.37ITKK302 pKa = 10.51AIYY305 pKa = 10.16EE306 pKa = 4.13NTEE309 pKa = 3.87NMSHH313 pKa = 6.53AKK315 pKa = 10.57AEE317 pKa = 4.62FIQVEE322 pKa = 4.27TALDD326 pKa = 4.51GIGIDD331 pKa = 3.64LHH333 pKa = 6.76PGAEE337 pKa = 4.26KK338 pKa = 10.85YY339 pKa = 8.47FTEE342 pKa = 5.04AGVLEE347 pKa = 4.37
Molecular weight: 36.15 kDa
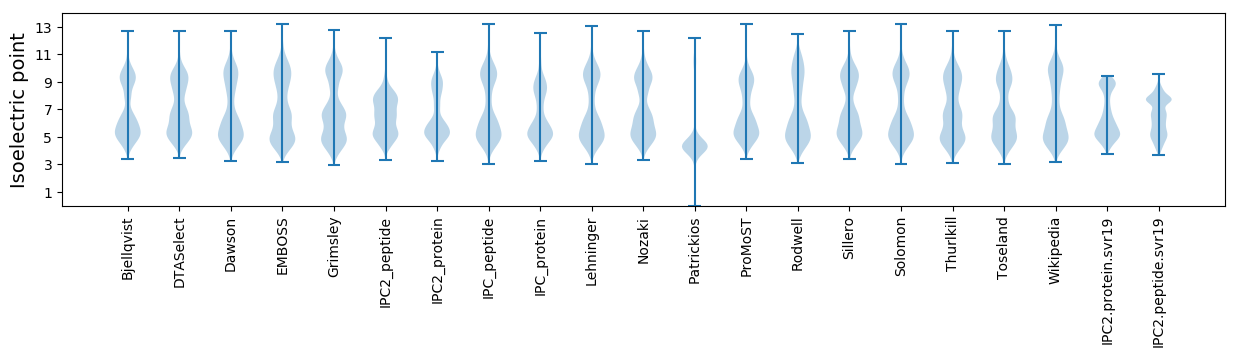
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1U9K5F7|A0A1U9K5F7_9BACL Uncharacterized protein OS=Novibacillus thermophilus OX=1471761 GN=B0W44_05315 PE=4 SV=1
MM1 pKa = 7.4NGRR4 pKa = 11.84RR5 pKa = 11.84MLWTTLIGAAITVIARR21 pKa = 11.84AKK23 pKa = 9.86RR24 pKa = 11.84HH25 pKa = 5.23RR26 pKa = 11.84QAPLARR32 pKa = 11.84LFRR35 pKa = 11.84GAGRR39 pKa = 11.84GNKK42 pKa = 7.74MLWMRR47 pKa = 11.84YY48 pKa = 4.68TQPVMHH54 pKa = 6.72MVRR57 pKa = 11.84RR58 pKa = 11.84MARR61 pKa = 3.08
MM1 pKa = 7.4NGRR4 pKa = 11.84RR5 pKa = 11.84MLWTTLIGAAITVIARR21 pKa = 11.84AKK23 pKa = 9.86RR24 pKa = 11.84HH25 pKa = 5.23RR26 pKa = 11.84QAPLARR32 pKa = 11.84LFRR35 pKa = 11.84GAGRR39 pKa = 11.84GNKK42 pKa = 7.74MLWMRR47 pKa = 11.84YY48 pKa = 4.68TQPVMHH54 pKa = 6.72MVRR57 pKa = 11.84RR58 pKa = 11.84MARR61 pKa = 3.08
Molecular weight: 7.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
951385 |
27 |
1459 |
300.2 |
33.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.918 ± 0.041 | 0.793 ± 0.013 |
5.445 ± 0.032 | 7.241 ± 0.05 |
4.111 ± 0.033 | 7.362 ± 0.04 |
2.362 ± 0.022 | 6.143 ± 0.038 |
5.253 ± 0.037 | 9.835 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.643 ± 0.019 | 3.386 ± 0.027 |
4.152 ± 0.028 | 3.74 ± 0.024 |
5.858 ± 0.045 | 5.493 ± 0.028 |
5.419 ± 0.028 | 8.307 ± 0.042 |
1.293 ± 0.019 | 3.247 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
