
Halobiforma nitratireducens JCM 10879
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Halobiforma; Halobiforma nitratireducens
Average proteome isoelectric point is 4.81
Get precalculated fractions of proteins
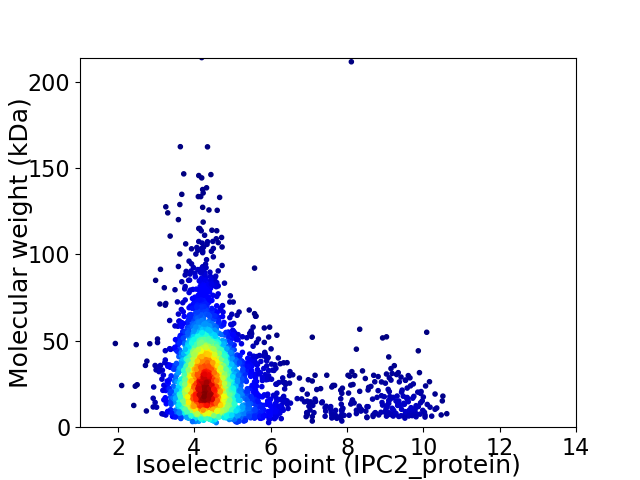
Virtual 2D-PAGE plot for 3534 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0LCS2|M0LCS2_9EURY Polysulfide reductase NrfD (Fragment) OS=Halobiforma nitratireducens JCM 10879 OX=1227454 GN=C446_15383 PE=4 SV=1
MM1 pKa = 6.54TASVALAGCIGGNGNGDD18 pKa = 3.9DD19 pKa = 4.55NGDD22 pKa = 3.74DD23 pKa = 3.51VADD26 pKa = 3.51ISEE29 pKa = 4.13YY30 pKa = 10.55QYY32 pKa = 11.69DD33 pKa = 3.85RR34 pKa = 11.84EE35 pKa = 4.25EE36 pKa = 4.81PDD38 pKa = 4.32DD39 pKa = 4.05EE40 pKa = 4.88DD41 pKa = 6.14RR42 pKa = 11.84SDD44 pKa = 3.78SLEE47 pKa = 3.73FLQPAEE53 pKa = 4.36RR54 pKa = 11.84DD55 pKa = 3.27EE56 pKa = 5.9DD57 pKa = 4.11FDD59 pKa = 4.38PVVSFDD65 pKa = 3.72SYY67 pKa = 11.19SAQVMNLVFDD77 pKa = 5.28NLYY80 pKa = 10.65EE81 pKa = 4.21WDD83 pKa = 4.53DD84 pKa = 3.82EE85 pKa = 4.44MGLEE89 pKa = 4.09PKK91 pKa = 10.05IADD94 pKa = 3.91GMPEE98 pKa = 3.96EE99 pKa = 4.23QDD101 pKa = 3.76DD102 pKa = 4.36GEE104 pKa = 4.71TYY106 pKa = 10.82VFEE109 pKa = 4.48IKK111 pKa = 10.46EE112 pKa = 4.38GIEE115 pKa = 3.77FHH117 pKa = 7.05NGDD120 pKa = 4.21EE121 pKa = 4.23VTAADD126 pKa = 3.78VAHH129 pKa = 6.86SFTAPVEE136 pKa = 4.23EE137 pKa = 4.5EE138 pKa = 3.83TDD140 pKa = 3.54NAASYY145 pKa = 11.83AMIEE149 pKa = 4.17STEE152 pKa = 4.13VIDD155 pKa = 5.15DD156 pKa = 3.56HH157 pKa = 5.6TLEE160 pKa = 4.42VNLEE164 pKa = 4.03HH165 pKa = 7.46PYY167 pKa = 11.31GPFTMVTMGVEE178 pKa = 4.24VVPEE182 pKa = 3.95DD183 pKa = 3.85VRR185 pKa = 11.84TDD187 pKa = 3.37DD188 pKa = 4.55RR189 pKa = 11.84DD190 pKa = 3.64AFNTDD195 pKa = 4.4PIGSGPFQFADD206 pKa = 4.48FQQGEE211 pKa = 4.52YY212 pKa = 10.85VEE214 pKa = 4.38LEE216 pKa = 3.77RR217 pKa = 11.84WDD219 pKa = 5.08DD220 pKa = 3.72YY221 pKa = 11.12WDD223 pKa = 3.88EE224 pKa = 4.03PHH226 pKa = 6.66PHH228 pKa = 6.73IEE230 pKa = 4.2EE231 pKa = 4.01IRR233 pKa = 11.84FEE235 pKa = 4.23AAPDD239 pKa = 3.67DD240 pKa = 4.51ANRR243 pKa = 11.84IAQILAGDD251 pKa = 3.91TDD253 pKa = 4.4VIDD256 pKa = 3.9TVPATEE262 pKa = 3.78WDD264 pKa = 3.98EE265 pKa = 4.79VEE267 pKa = 4.39GADD270 pKa = 4.76DD271 pKa = 4.07VVIHH275 pKa = 6.31GSRR278 pKa = 11.84SPSYY282 pKa = 9.87MYY284 pKa = 10.97LAFNCNEE291 pKa = 4.16GEE293 pKa = 4.33TTNPDD298 pKa = 2.58VRR300 pKa = 11.84RR301 pKa = 11.84AVGHH305 pKa = 5.93SFSMHH310 pKa = 6.6DD311 pKa = 3.96FVNEE315 pKa = 4.01HH316 pKa = 7.03LGPAADD322 pKa = 4.13PLVSPVPEE330 pKa = 3.97ITNDD334 pKa = 3.27SGDD337 pKa = 3.14WDD339 pKa = 4.36FPVDD343 pKa = 3.73DD344 pKa = 4.53WAEE347 pKa = 3.94MMPEE351 pKa = 4.0YY352 pKa = 10.95DD353 pKa = 3.51PDD355 pKa = 3.46QAQEE359 pKa = 4.08LLEE362 pKa = 4.52DD363 pKa = 3.96AGVPEE368 pKa = 4.78DD369 pKa = 3.91WEE371 pKa = 4.19PTIIAPEE378 pKa = 4.35GGPRR382 pKa = 11.84EE383 pKa = 4.1ALAEE387 pKa = 4.42RR388 pKa = 11.84IGSRR392 pKa = 11.84LTEE395 pKa = 3.74IGYY398 pKa = 9.32GADD401 pKa = 3.41VQGMSFATLVDD412 pKa = 4.09TYY414 pKa = 9.2TTGNADD420 pKa = 3.84DD421 pKa = 4.4YY422 pKa = 11.96EE423 pKa = 4.43MYY425 pKa = 11.09LLGWTGGPDD434 pKa = 3.29PDD436 pKa = 3.86AYY438 pKa = 10.57FYY440 pKa = 11.64NLFHH444 pKa = 7.18EE445 pKa = 4.69SQEE448 pKa = 4.47GVGQGHH454 pKa = 6.76FYY456 pKa = 10.71EE457 pKa = 4.91GQGDD461 pKa = 3.89FHH463 pKa = 8.55DD464 pKa = 5.14SIMNARR470 pKa = 11.84QSADD474 pKa = 3.12QDD476 pKa = 3.68DD477 pKa = 3.95RR478 pKa = 11.84RR479 pKa = 11.84EE480 pKa = 3.99LYY482 pKa = 10.06IDD484 pKa = 3.4VTEE487 pKa = 5.15EE488 pKa = 3.54ILDD491 pKa = 3.84YY492 pKa = 10.9MPVLPAYY499 pKa = 10.22SEE501 pKa = 4.58HH502 pKa = 5.6NTMAARR508 pKa = 11.84EE509 pKa = 4.07EE510 pKa = 4.4VKK512 pKa = 10.63DD513 pKa = 3.85LHH515 pKa = 6.5AHH517 pKa = 6.27PAVSFNPRR525 pKa = 11.84IVSNYY530 pKa = 7.25QNTWIDD536 pKa = 3.42EE537 pKa = 4.24
MM1 pKa = 6.54TASVALAGCIGGNGNGDD18 pKa = 3.9DD19 pKa = 4.55NGDD22 pKa = 3.74DD23 pKa = 3.51VADD26 pKa = 3.51ISEE29 pKa = 4.13YY30 pKa = 10.55QYY32 pKa = 11.69DD33 pKa = 3.85RR34 pKa = 11.84EE35 pKa = 4.25EE36 pKa = 4.81PDD38 pKa = 4.32DD39 pKa = 4.05EE40 pKa = 4.88DD41 pKa = 6.14RR42 pKa = 11.84SDD44 pKa = 3.78SLEE47 pKa = 3.73FLQPAEE53 pKa = 4.36RR54 pKa = 11.84DD55 pKa = 3.27EE56 pKa = 5.9DD57 pKa = 4.11FDD59 pKa = 4.38PVVSFDD65 pKa = 3.72SYY67 pKa = 11.19SAQVMNLVFDD77 pKa = 5.28NLYY80 pKa = 10.65EE81 pKa = 4.21WDD83 pKa = 4.53DD84 pKa = 3.82EE85 pKa = 4.44MGLEE89 pKa = 4.09PKK91 pKa = 10.05IADD94 pKa = 3.91GMPEE98 pKa = 3.96EE99 pKa = 4.23QDD101 pKa = 3.76DD102 pKa = 4.36GEE104 pKa = 4.71TYY106 pKa = 10.82VFEE109 pKa = 4.48IKK111 pKa = 10.46EE112 pKa = 4.38GIEE115 pKa = 3.77FHH117 pKa = 7.05NGDD120 pKa = 4.21EE121 pKa = 4.23VTAADD126 pKa = 3.78VAHH129 pKa = 6.86SFTAPVEE136 pKa = 4.23EE137 pKa = 4.5EE138 pKa = 3.83TDD140 pKa = 3.54NAASYY145 pKa = 11.83AMIEE149 pKa = 4.17STEE152 pKa = 4.13VIDD155 pKa = 5.15DD156 pKa = 3.56HH157 pKa = 5.6TLEE160 pKa = 4.42VNLEE164 pKa = 4.03HH165 pKa = 7.46PYY167 pKa = 11.31GPFTMVTMGVEE178 pKa = 4.24VVPEE182 pKa = 3.95DD183 pKa = 3.85VRR185 pKa = 11.84TDD187 pKa = 3.37DD188 pKa = 4.55RR189 pKa = 11.84DD190 pKa = 3.64AFNTDD195 pKa = 4.4PIGSGPFQFADD206 pKa = 4.48FQQGEE211 pKa = 4.52YY212 pKa = 10.85VEE214 pKa = 4.38LEE216 pKa = 3.77RR217 pKa = 11.84WDD219 pKa = 5.08DD220 pKa = 3.72YY221 pKa = 11.12WDD223 pKa = 3.88EE224 pKa = 4.03PHH226 pKa = 6.66PHH228 pKa = 6.73IEE230 pKa = 4.2EE231 pKa = 4.01IRR233 pKa = 11.84FEE235 pKa = 4.23AAPDD239 pKa = 3.67DD240 pKa = 4.51ANRR243 pKa = 11.84IAQILAGDD251 pKa = 3.91TDD253 pKa = 4.4VIDD256 pKa = 3.9TVPATEE262 pKa = 3.78WDD264 pKa = 3.98EE265 pKa = 4.79VEE267 pKa = 4.39GADD270 pKa = 4.76DD271 pKa = 4.07VVIHH275 pKa = 6.31GSRR278 pKa = 11.84SPSYY282 pKa = 9.87MYY284 pKa = 10.97LAFNCNEE291 pKa = 4.16GEE293 pKa = 4.33TTNPDD298 pKa = 2.58VRR300 pKa = 11.84RR301 pKa = 11.84AVGHH305 pKa = 5.93SFSMHH310 pKa = 6.6DD311 pKa = 3.96FVNEE315 pKa = 4.01HH316 pKa = 7.03LGPAADD322 pKa = 4.13PLVSPVPEE330 pKa = 3.97ITNDD334 pKa = 3.27SGDD337 pKa = 3.14WDD339 pKa = 4.36FPVDD343 pKa = 3.73DD344 pKa = 4.53WAEE347 pKa = 3.94MMPEE351 pKa = 4.0YY352 pKa = 10.95DD353 pKa = 3.51PDD355 pKa = 3.46QAQEE359 pKa = 4.08LLEE362 pKa = 4.52DD363 pKa = 3.96AGVPEE368 pKa = 4.78DD369 pKa = 3.91WEE371 pKa = 4.19PTIIAPEE378 pKa = 4.35GGPRR382 pKa = 11.84EE383 pKa = 4.1ALAEE387 pKa = 4.42RR388 pKa = 11.84IGSRR392 pKa = 11.84LTEE395 pKa = 3.74IGYY398 pKa = 9.32GADD401 pKa = 3.41VQGMSFATLVDD412 pKa = 4.09TYY414 pKa = 9.2TTGNADD420 pKa = 3.84DD421 pKa = 4.4YY422 pKa = 11.96EE423 pKa = 4.43MYY425 pKa = 11.09LLGWTGGPDD434 pKa = 3.29PDD436 pKa = 3.86AYY438 pKa = 10.57FYY440 pKa = 11.64NLFHH444 pKa = 7.18EE445 pKa = 4.69SQEE448 pKa = 4.47GVGQGHH454 pKa = 6.76FYY456 pKa = 10.71EE457 pKa = 4.91GQGDD461 pKa = 3.89FHH463 pKa = 8.55DD464 pKa = 5.14SIMNARR470 pKa = 11.84QSADD474 pKa = 3.12QDD476 pKa = 3.68DD477 pKa = 3.95RR478 pKa = 11.84RR479 pKa = 11.84EE480 pKa = 3.99LYY482 pKa = 10.06IDD484 pKa = 3.4VTEE487 pKa = 5.15EE488 pKa = 3.54ILDD491 pKa = 3.84YY492 pKa = 10.9MPVLPAYY499 pKa = 10.22SEE501 pKa = 4.58HH502 pKa = 5.6NTMAARR508 pKa = 11.84EE509 pKa = 4.07EE510 pKa = 4.4VKK512 pKa = 10.63DD513 pKa = 3.85LHH515 pKa = 6.5AHH517 pKa = 6.27PAVSFNPRR525 pKa = 11.84IVSNYY530 pKa = 7.25QNTWIDD536 pKa = 3.42EE537 pKa = 4.24
Molecular weight: 60.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0MRF1|M0MRF1_9EURY 50S ribosomal protein L4 OS=Halobiforma nitratireducens JCM 10879 OX=1227454 GN=rpl4lp PE=3 SV=1
MM1 pKa = 7.69TGTQTRR7 pKa = 11.84ATALLEE13 pKa = 4.11PVRR16 pKa = 11.84DD17 pKa = 3.59RR18 pKa = 11.84VPVVLVPVLLGGVLLLYY35 pKa = 9.69FVVPLAAFFLQTGTVDD51 pKa = 3.68VIGGLGDD58 pKa = 3.69PVTRR62 pKa = 11.84DD63 pKa = 3.8AIATSLLTAPVATAISTVFGVPLAYY88 pKa = 10.34VLSRR92 pKa = 11.84FSFRR96 pKa = 11.84GKK98 pKa = 10.18RR99 pKa = 11.84LVLAAVLVPLVLPPVVGGAVLLSVLGQFTPIGAAAADD136 pKa = 3.8AGVPLTDD143 pKa = 3.13SRR145 pKa = 11.84LGIVLAQTFVAAPFLVITAKK165 pKa = 10.8AGFDD169 pKa = 3.6DD170 pKa = 3.68VDD172 pKa = 3.59PRR174 pKa = 11.84LEE176 pKa = 3.87EE177 pKa = 3.86AARR180 pKa = 11.84TMGYY184 pKa = 10.79GPIATVRR191 pKa = 11.84RR192 pKa = 11.84VSLPLARR199 pKa = 11.84NAIAAGIVLTFVRR212 pKa = 11.84AIGEE216 pKa = 4.27FGATMMVAYY225 pKa = 10.14NPRR228 pKa = 11.84TMPTRR233 pKa = 11.84IWVAFVSRR241 pKa = 11.84GLEE244 pKa = 3.64AVVPIVVALLVITILVVVAVQLLVGTPKK272 pKa = 10.63RR273 pKa = 11.84FF274 pKa = 3.18
MM1 pKa = 7.69TGTQTRR7 pKa = 11.84ATALLEE13 pKa = 4.11PVRR16 pKa = 11.84DD17 pKa = 3.59RR18 pKa = 11.84VPVVLVPVLLGGVLLLYY35 pKa = 9.69FVVPLAAFFLQTGTVDD51 pKa = 3.68VIGGLGDD58 pKa = 3.69PVTRR62 pKa = 11.84DD63 pKa = 3.8AIATSLLTAPVATAISTVFGVPLAYY88 pKa = 10.34VLSRR92 pKa = 11.84FSFRR96 pKa = 11.84GKK98 pKa = 10.18RR99 pKa = 11.84LVLAAVLVPLVLPPVVGGAVLLSVLGQFTPIGAAAADD136 pKa = 3.8AGVPLTDD143 pKa = 3.13SRR145 pKa = 11.84LGIVLAQTFVAAPFLVITAKK165 pKa = 10.8AGFDD169 pKa = 3.6DD170 pKa = 3.68VDD172 pKa = 3.59PRR174 pKa = 11.84LEE176 pKa = 3.87EE177 pKa = 3.86AARR180 pKa = 11.84TMGYY184 pKa = 10.79GPIATVRR191 pKa = 11.84RR192 pKa = 11.84VSLPLARR199 pKa = 11.84NAIAAGIVLTFVRR212 pKa = 11.84AIGEE216 pKa = 4.27FGATMMVAYY225 pKa = 10.14NPRR228 pKa = 11.84TMPTRR233 pKa = 11.84IWVAFVSRR241 pKa = 11.84GLEE244 pKa = 3.64AVVPIVVALLVITILVVVAVQLLVGTPKK272 pKa = 10.63RR273 pKa = 11.84FF274 pKa = 3.18
Molecular weight: 28.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1004006 |
24 |
2023 |
284.1 |
30.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.431 ± 0.047 | 0.739 ± 0.014 |
9.03 ± 0.057 | 9.542 ± 0.067 |
3.14 ± 0.029 | 8.448 ± 0.048 |
1.99 ± 0.02 | 4.101 ± 0.031 |
1.625 ± 0.025 | 8.746 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.63 ± 0.019 | 2.171 ± 0.021 |
4.75 ± 0.027 | 2.33 ± 0.024 |
6.667 ± 0.054 | 5.542 ± 0.042 |
6.481 ± 0.034 | 8.882 ± 0.048 |
1.109 ± 0.018 | 2.642 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
