
Rhizobacter sp. Root404
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiales genera incertae sedis; Rhizobacter; unclassified Rhizobacter
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
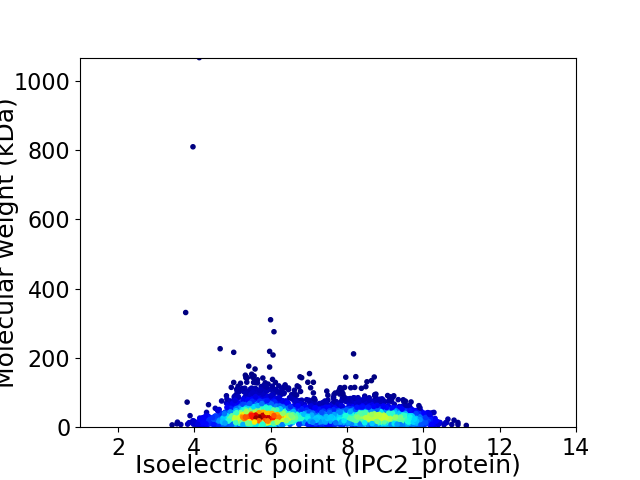
Virtual 2D-PAGE plot for 4707 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7AGQ4|A0A0Q7AGQ4_9BURK Guanylate cyclase domain-containing protein OS=Rhizobacter sp. Root404 OX=1736528 GN=ASC76_22885 PE=4 SV=1
MM1 pKa = 6.2TTEE4 pKa = 4.08AVALASYY11 pKa = 9.52SVKK14 pKa = 9.21ITSLPVLGDD23 pKa = 3.34LMYY26 pKa = 10.94FNGVSLVPVALNSTYY41 pKa = 10.43TYY43 pKa = 11.47ADD45 pKa = 3.81IIAGKK50 pKa = 9.47LQFVPDD56 pKa = 4.34ANQSGIDD63 pKa = 3.82GYY65 pKa = 11.43GAAGTGNKK73 pKa = 10.08LNDD76 pKa = 3.34YY77 pKa = 10.94ASFNFEE83 pKa = 4.28VIANGINSNPATVVIDD99 pKa = 3.69IAPVADD105 pKa = 4.7APTLSLTTASFLRR118 pKa = 11.84TSSLDD123 pKa = 3.23EE124 pKa = 4.06LTAMSGPNAWTRR136 pKa = 11.84YY137 pKa = 6.85ATDD140 pKa = 3.47MNGGAWRR147 pKa = 11.84TSNSGGTIEE156 pKa = 4.72VYY158 pKa = 11.14ADD160 pKa = 3.74GEE162 pKa = 4.33YY163 pKa = 10.63LQNGGSNRR171 pKa = 11.84VIEE174 pKa = 4.51LEE176 pKa = 4.32SNTGAAADD184 pKa = 5.07LYY186 pKa = 10.93TDD188 pKa = 3.72VATKK192 pKa = 10.47AGQVYY197 pKa = 10.23HH198 pKa = 6.93LSLDD202 pKa = 3.47FAEE205 pKa = 4.63RR206 pKa = 11.84LNNQSMASQGLIPAYY221 pKa = 9.02GTAQIDD227 pKa = 4.62VYY229 pKa = 10.45WGGVKK234 pKa = 9.93VATLNTDD241 pKa = 3.17TSTWTHH247 pKa = 6.38FEE249 pKa = 4.15LDD251 pKa = 3.46LAATTTGTTRR261 pKa = 11.84LTFQATDD268 pKa = 3.55SNSLGGVMDD277 pKa = 4.27NLSLQLSQNTTMRR290 pKa = 11.84NTTTQLPQPTIALVDD305 pKa = 3.68TDD307 pKa = 3.82GSEE310 pKa = 4.19TLALTVTGVPIGATISDD327 pKa = 3.89GTRR330 pKa = 11.84SFTATAGNQVATITGWTYY348 pKa = 11.19SALTVTPPAGYY359 pKa = 8.53TGSFTLNYY367 pKa = 7.78TATSTEE373 pKa = 3.93AVGGSTATTITPVTVNVIAPNTGPVATNDD402 pKa = 3.48TVTGIRR408 pKa = 11.84DD409 pKa = 3.67VVVHH413 pKa = 6.35GNVLTNDD420 pKa = 3.11TDD422 pKa = 4.31ADD424 pKa = 4.18GNTLYY429 pKa = 9.77VTGFTVNGTNYY440 pKa = 8.88ATYY443 pKa = 10.69DD444 pKa = 3.72SVGVTDD450 pKa = 3.64NTTDD454 pKa = 3.02NVTIAGVGYY463 pKa = 10.16FRR465 pKa = 11.84MEE467 pKa = 4.02ADD469 pKa = 3.08GSYY472 pKa = 10.5QFTPLAGYY480 pKa = 8.04TGTAPAITYY489 pKa = 7.16TTADD493 pKa = 3.17EE494 pKa = 4.57HH495 pKa = 7.06GGYY498 pKa = 7.91STATLTLTEE507 pKa = 4.42TTTGGIIGTSSAEE520 pKa = 4.27TLNGTAGADD529 pKa = 3.65VISASGGIDD538 pKa = 3.26TLNGLGGNDD547 pKa = 3.72VLDD550 pKa = 4.54GGAGADD556 pKa = 3.62VLVGGAGSDD565 pKa = 3.57VLYY568 pKa = 11.08GRR570 pKa = 11.84AGNDD574 pKa = 3.09TLTGGTGTGTDD585 pKa = 3.32TASNTFAWAFGDD597 pKa = 3.54QGTTSTLAVDD607 pKa = 4.91TITDD611 pKa = 3.89FNKK614 pKa = 10.34AAASSGGDD622 pKa = 3.43VLDD625 pKa = 5.63LRR627 pKa = 11.84DD628 pKa = 3.92LLIGEE633 pKa = 4.36YY634 pKa = 10.64HH635 pKa = 7.11NAFQPNGNLTDD646 pKa = 4.07FLHH649 pKa = 6.31FTTSAGTTTIEE660 pKa = 4.42VKK662 pKa = 10.31SHH664 pKa = 5.46GVGTSSADD672 pKa = 3.42EE673 pKa = 4.76KK674 pKa = 11.01IVLTGVDD681 pKa = 4.11LTNGGALNTDD691 pKa = 3.17QLIIQDD697 pKa = 4.66LLSKK701 pKa = 10.89GKK703 pKa = 10.45LVVDD707 pKa = 4.31
MM1 pKa = 6.2TTEE4 pKa = 4.08AVALASYY11 pKa = 9.52SVKK14 pKa = 9.21ITSLPVLGDD23 pKa = 3.34LMYY26 pKa = 10.94FNGVSLVPVALNSTYY41 pKa = 10.43TYY43 pKa = 11.47ADD45 pKa = 3.81IIAGKK50 pKa = 9.47LQFVPDD56 pKa = 4.34ANQSGIDD63 pKa = 3.82GYY65 pKa = 11.43GAAGTGNKK73 pKa = 10.08LNDD76 pKa = 3.34YY77 pKa = 10.94ASFNFEE83 pKa = 4.28VIANGINSNPATVVIDD99 pKa = 3.69IAPVADD105 pKa = 4.7APTLSLTTASFLRR118 pKa = 11.84TSSLDD123 pKa = 3.23EE124 pKa = 4.06LTAMSGPNAWTRR136 pKa = 11.84YY137 pKa = 6.85ATDD140 pKa = 3.47MNGGAWRR147 pKa = 11.84TSNSGGTIEE156 pKa = 4.72VYY158 pKa = 11.14ADD160 pKa = 3.74GEE162 pKa = 4.33YY163 pKa = 10.63LQNGGSNRR171 pKa = 11.84VIEE174 pKa = 4.51LEE176 pKa = 4.32SNTGAAADD184 pKa = 5.07LYY186 pKa = 10.93TDD188 pKa = 3.72VATKK192 pKa = 10.47AGQVYY197 pKa = 10.23HH198 pKa = 6.93LSLDD202 pKa = 3.47FAEE205 pKa = 4.63RR206 pKa = 11.84LNNQSMASQGLIPAYY221 pKa = 9.02GTAQIDD227 pKa = 4.62VYY229 pKa = 10.45WGGVKK234 pKa = 9.93VATLNTDD241 pKa = 3.17TSTWTHH247 pKa = 6.38FEE249 pKa = 4.15LDD251 pKa = 3.46LAATTTGTTRR261 pKa = 11.84LTFQATDD268 pKa = 3.55SNSLGGVMDD277 pKa = 4.27NLSLQLSQNTTMRR290 pKa = 11.84NTTTQLPQPTIALVDD305 pKa = 3.68TDD307 pKa = 3.82GSEE310 pKa = 4.19TLALTVTGVPIGATISDD327 pKa = 3.89GTRR330 pKa = 11.84SFTATAGNQVATITGWTYY348 pKa = 11.19SALTVTPPAGYY359 pKa = 8.53TGSFTLNYY367 pKa = 7.78TATSTEE373 pKa = 3.93AVGGSTATTITPVTVNVIAPNTGPVATNDD402 pKa = 3.48TVTGIRR408 pKa = 11.84DD409 pKa = 3.67VVVHH413 pKa = 6.35GNVLTNDD420 pKa = 3.11TDD422 pKa = 4.31ADD424 pKa = 4.18GNTLYY429 pKa = 9.77VTGFTVNGTNYY440 pKa = 8.88ATYY443 pKa = 10.69DD444 pKa = 3.72SVGVTDD450 pKa = 3.64NTTDD454 pKa = 3.02NVTIAGVGYY463 pKa = 10.16FRR465 pKa = 11.84MEE467 pKa = 4.02ADD469 pKa = 3.08GSYY472 pKa = 10.5QFTPLAGYY480 pKa = 8.04TGTAPAITYY489 pKa = 7.16TTADD493 pKa = 3.17EE494 pKa = 4.57HH495 pKa = 7.06GGYY498 pKa = 7.91STATLTLTEE507 pKa = 4.42TTTGGIIGTSSAEE520 pKa = 4.27TLNGTAGADD529 pKa = 3.65VISASGGIDD538 pKa = 3.26TLNGLGGNDD547 pKa = 3.72VLDD550 pKa = 4.54GGAGADD556 pKa = 3.62VLVGGAGSDD565 pKa = 3.57VLYY568 pKa = 11.08GRR570 pKa = 11.84AGNDD574 pKa = 3.09TLTGGTGTGTDD585 pKa = 3.32TASNTFAWAFGDD597 pKa = 3.54QGTTSTLAVDD607 pKa = 4.91TITDD611 pKa = 3.89FNKK614 pKa = 10.34AAASSGGDD622 pKa = 3.43VLDD625 pKa = 5.63LRR627 pKa = 11.84DD628 pKa = 3.92LLIGEE633 pKa = 4.36YY634 pKa = 10.64HH635 pKa = 7.11NAFQPNGNLTDD646 pKa = 4.07FLHH649 pKa = 6.31FTTSAGTTTIEE660 pKa = 4.42VKK662 pKa = 10.31SHH664 pKa = 5.46GVGTSSADD672 pKa = 3.42EE673 pKa = 4.76KK674 pKa = 11.01IVLTGVDD681 pKa = 4.11LTNGGALNTDD691 pKa = 3.17QLIIQDD697 pKa = 4.66LLSKK701 pKa = 10.89GKK703 pKa = 10.45LVVDD707 pKa = 4.31
Molecular weight: 72.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7AKX4|A0A0Q7AKX4_9BURK Nicotinate phosphoribosyltransferase OS=Rhizobacter sp. Root404 OX=1736528 GN=ASC76_06030 PE=3 SV=1
MM1 pKa = 7.33KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.71QASKK9 pKa = 9.04VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.33KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 9.71QASKK9 pKa = 9.04VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1567174 |
44 |
10585 |
332.9 |
35.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.683 ± 0.06 | 0.86 ± 0.012 |
5.454 ± 0.039 | 4.997 ± 0.037 |
3.507 ± 0.024 | 8.45 ± 0.048 |
2.196 ± 0.02 | 4.253 ± 0.023 |
3.014 ± 0.034 | 10.683 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.384 ± 0.022 | 2.462 ± 0.026 |
5.348 ± 0.029 | 3.533 ± 0.02 |
7.205 ± 0.046 | 5.133 ± 0.03 |
5.407 ± 0.041 | 7.893 ± 0.029 |
1.456 ± 0.017 | 2.083 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
