
Flavimaricola marinus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Flavimaricola
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
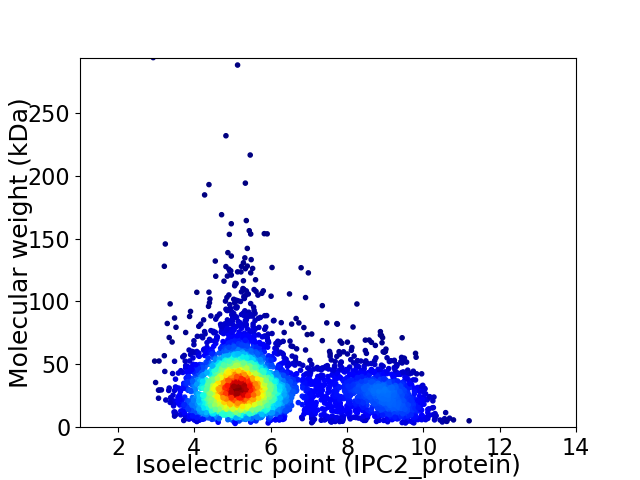
Virtual 2D-PAGE plot for 4548 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238LFH6|A0A238LFH6_9RHOB Carnitinyl-CoA dehydratase OS=Flavimaricola marinus OX=1819565 GN=caiD_1 PE=3 SV=1
MM1 pKa = 6.2VTGVEE6 pKa = 4.07LTYY9 pKa = 10.11DD10 pKa = 3.58TGADD14 pKa = 3.31AMEE17 pKa = 4.09MAEE20 pKa = 5.06AIFGDD25 pKa = 3.99SVEE28 pKa = 4.53VVSASYY34 pKa = 10.47SGSYY38 pKa = 9.69YY39 pKa = 10.84SSAIYY44 pKa = 10.3TNGEE48 pKa = 4.29SISPNVVPGDD58 pKa = 3.5TGVILSTGAATYY70 pKa = 7.5FTNSGGGSNDD80 pKa = 3.47YY81 pKa = 11.54SNTSYY86 pKa = 11.61NSGGEE91 pKa = 4.09NYY93 pKa = 10.13NDD95 pKa = 4.0SFDD98 pKa = 4.93DD99 pKa = 3.64VAGTYY104 pKa = 9.47TYY106 pKa = 11.03DD107 pKa = 3.29AAYY110 pKa = 9.01MDD112 pKa = 4.22IVFIPDD118 pKa = 3.38GDD120 pKa = 4.02VMTMQFVFASDD131 pKa = 4.29EE132 pKa = 4.12YY133 pKa = 10.77PEE135 pKa = 4.49YY136 pKa = 10.88SDD138 pKa = 5.55SIYY141 pKa = 11.06NDD143 pKa = 3.16VVLVEE148 pKa = 4.42VNGEE152 pKa = 4.19VIPLSVTNSSTSVTEE167 pKa = 3.7INQNEE172 pKa = 4.43NVNLYY177 pKa = 10.5NDD179 pKa = 3.84NTADD183 pKa = 3.52QYY185 pKa = 9.5NTEE188 pKa = 4.03MDD190 pKa = 3.79GFTVTMTLTIPVVAGEE206 pKa = 4.24EE207 pKa = 3.74NSIRR211 pKa = 11.84IGIADD216 pKa = 4.28ASDD219 pKa = 3.49SNYY222 pKa = 10.97DD223 pKa = 3.68SNLLIAGDD231 pKa = 4.04SLQTDD236 pKa = 4.22LVAIQDD242 pKa = 3.86DD243 pKa = 4.39VTVKK247 pKa = 10.76EE248 pKa = 4.77GVTKK252 pKa = 10.21TVDD255 pKa = 3.72VLDD258 pKa = 4.24NDD260 pKa = 4.48LNATSGTLVITHH272 pKa = 6.87INDD275 pKa = 3.18IAVTAGDD282 pKa = 4.21TVNLTTGQSVTLNADD297 pKa = 3.25GTLSITADD305 pKa = 3.47SDD307 pKa = 3.54NDD309 pKa = 4.23TVSFTYY315 pKa = 8.58EE316 pKa = 3.45TAAEE320 pKa = 4.51DD321 pKa = 3.81GSGAVLEE328 pKa = 4.34TDD330 pKa = 3.42VGFVTVTTVPCFVAGTRR347 pKa = 11.84ISTPDD352 pKa = 2.93GDD354 pKa = 4.44RR355 pKa = 11.84RR356 pKa = 11.84VEE358 pKa = 3.96DD359 pKa = 4.34LQPGDD364 pKa = 4.21LVLTKK369 pKa = 10.57DD370 pKa = 3.69AGAQQVRR377 pKa = 11.84WAGRR381 pKa = 11.84RR382 pKa = 11.84SVAAQGRR389 pKa = 11.84MAPVLIRR396 pKa = 11.84SGTFGTHH403 pKa = 5.6GDD405 pKa = 3.72VMVSPLHH412 pKa = 6.03RR413 pKa = 11.84VLVTGGVAEE422 pKa = 4.61LLFAEE427 pKa = 4.68PEE429 pKa = 4.37VLVTARR435 pKa = 11.84DD436 pKa = 3.73LVNDD440 pKa = 3.94RR441 pKa = 11.84SVRR444 pKa = 11.84RR445 pKa = 11.84VEE447 pKa = 4.45GGTVDD452 pKa = 3.41YY453 pKa = 10.72VHH455 pKa = 8.03IMFDD459 pKa = 3.18AHH461 pKa = 5.35QVIYY465 pKa = 10.9ADD467 pKa = 3.95GLATEE472 pKa = 4.9SFLPGPQTTSLFEE485 pKa = 4.27DD486 pKa = 4.27EE487 pKa = 5.34IVAEE491 pKa = 4.16ICAIFPEE498 pKa = 4.74LDD500 pKa = 3.33PVTGEE505 pKa = 4.56GYY507 pKa = 10.4SPAARR512 pKa = 11.84RR513 pKa = 11.84TLRR516 pKa = 11.84AYY518 pKa = 9.73EE519 pKa = 4.08GRR521 pKa = 11.84LLGQVAA527 pKa = 3.83
MM1 pKa = 6.2VTGVEE6 pKa = 4.07LTYY9 pKa = 10.11DD10 pKa = 3.58TGADD14 pKa = 3.31AMEE17 pKa = 4.09MAEE20 pKa = 5.06AIFGDD25 pKa = 3.99SVEE28 pKa = 4.53VVSASYY34 pKa = 10.47SGSYY38 pKa = 9.69YY39 pKa = 10.84SSAIYY44 pKa = 10.3TNGEE48 pKa = 4.29SISPNVVPGDD58 pKa = 3.5TGVILSTGAATYY70 pKa = 7.5FTNSGGGSNDD80 pKa = 3.47YY81 pKa = 11.54SNTSYY86 pKa = 11.61NSGGEE91 pKa = 4.09NYY93 pKa = 10.13NDD95 pKa = 4.0SFDD98 pKa = 4.93DD99 pKa = 3.64VAGTYY104 pKa = 9.47TYY106 pKa = 11.03DD107 pKa = 3.29AAYY110 pKa = 9.01MDD112 pKa = 4.22IVFIPDD118 pKa = 3.38GDD120 pKa = 4.02VMTMQFVFASDD131 pKa = 4.29EE132 pKa = 4.12YY133 pKa = 10.77PEE135 pKa = 4.49YY136 pKa = 10.88SDD138 pKa = 5.55SIYY141 pKa = 11.06NDD143 pKa = 3.16VVLVEE148 pKa = 4.42VNGEE152 pKa = 4.19VIPLSVTNSSTSVTEE167 pKa = 3.7INQNEE172 pKa = 4.43NVNLYY177 pKa = 10.5NDD179 pKa = 3.84NTADD183 pKa = 3.52QYY185 pKa = 9.5NTEE188 pKa = 4.03MDD190 pKa = 3.79GFTVTMTLTIPVVAGEE206 pKa = 4.24EE207 pKa = 3.74NSIRR211 pKa = 11.84IGIADD216 pKa = 4.28ASDD219 pKa = 3.49SNYY222 pKa = 10.97DD223 pKa = 3.68SNLLIAGDD231 pKa = 4.04SLQTDD236 pKa = 4.22LVAIQDD242 pKa = 3.86DD243 pKa = 4.39VTVKK247 pKa = 10.76EE248 pKa = 4.77GVTKK252 pKa = 10.21TVDD255 pKa = 3.72VLDD258 pKa = 4.24NDD260 pKa = 4.48LNATSGTLVITHH272 pKa = 6.87INDD275 pKa = 3.18IAVTAGDD282 pKa = 4.21TVNLTTGQSVTLNADD297 pKa = 3.25GTLSITADD305 pKa = 3.47SDD307 pKa = 3.54NDD309 pKa = 4.23TVSFTYY315 pKa = 8.58EE316 pKa = 3.45TAAEE320 pKa = 4.51DD321 pKa = 3.81GSGAVLEE328 pKa = 4.34TDD330 pKa = 3.42VGFVTVTTVPCFVAGTRR347 pKa = 11.84ISTPDD352 pKa = 2.93GDD354 pKa = 4.44RR355 pKa = 11.84RR356 pKa = 11.84VEE358 pKa = 3.96DD359 pKa = 4.34LQPGDD364 pKa = 4.21LVLTKK369 pKa = 10.57DD370 pKa = 3.69AGAQQVRR377 pKa = 11.84WAGRR381 pKa = 11.84RR382 pKa = 11.84SVAAQGRR389 pKa = 11.84MAPVLIRR396 pKa = 11.84SGTFGTHH403 pKa = 5.6GDD405 pKa = 3.72VMVSPLHH412 pKa = 6.03RR413 pKa = 11.84VLVTGGVAEE422 pKa = 4.61LLFAEE427 pKa = 4.68PEE429 pKa = 4.37VLVTARR435 pKa = 11.84DD436 pKa = 3.73LVNDD440 pKa = 3.94RR441 pKa = 11.84SVRR444 pKa = 11.84RR445 pKa = 11.84VEE447 pKa = 4.45GGTVDD452 pKa = 3.41YY453 pKa = 10.72VHH455 pKa = 8.03IMFDD459 pKa = 3.18AHH461 pKa = 5.35QVIYY465 pKa = 10.9ADD467 pKa = 3.95GLATEE472 pKa = 4.9SFLPGPQTTSLFEE485 pKa = 4.27DD486 pKa = 4.27EE487 pKa = 5.34IVAEE491 pKa = 4.16ICAIFPEE498 pKa = 4.74LDD500 pKa = 3.33PVTGEE505 pKa = 4.56GYY507 pKa = 10.4SPAARR512 pKa = 11.84RR513 pKa = 11.84TLRR516 pKa = 11.84AYY518 pKa = 9.73EE519 pKa = 4.08GRR521 pKa = 11.84LLGQVAA527 pKa = 3.83
Molecular weight: 55.93 kDa
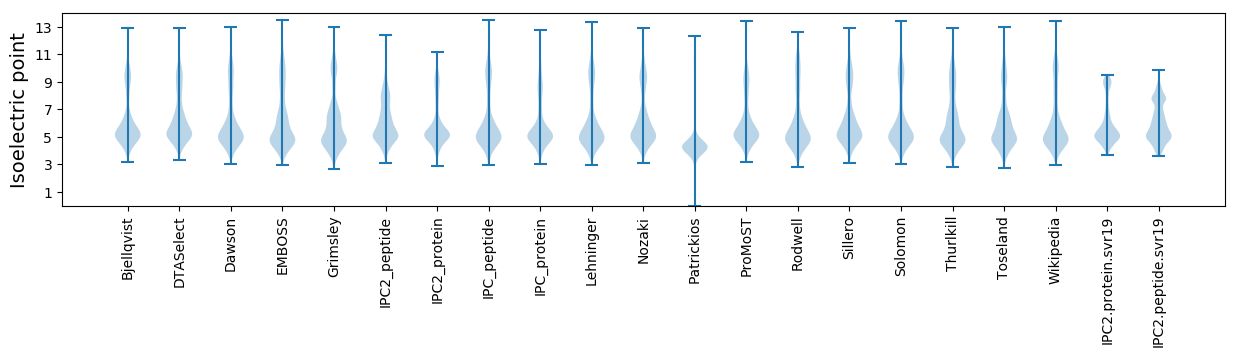
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238L9L3|A0A238L9L3_9RHOB Uncharacterized protein OS=Flavimaricola marinus OX=1819565 GN=LOM8899_00414 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.05ARR15 pKa = 11.84HH16 pKa = 4.79GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84SKK41 pKa = 11.13LSAA44 pKa = 3.7
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.05ARR15 pKa = 11.84HH16 pKa = 4.79GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.45AGRR28 pKa = 11.84KK29 pKa = 8.81IINARR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84SKK41 pKa = 11.13LSAA44 pKa = 3.7
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1434317 |
29 |
2947 |
315.4 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.253 ± 0.057 | 0.896 ± 0.01 |
6.228 ± 0.039 | 5.747 ± 0.033 |
3.709 ± 0.025 | 8.694 ± 0.038 |
2.037 ± 0.017 | 5.323 ± 0.027 |
2.903 ± 0.031 | 9.93 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.818 ± 0.017 | 2.582 ± 0.019 |
5.222 ± 0.028 | 3.178 ± 0.018 |
6.489 ± 0.038 | 5.344 ± 0.029 |
5.643 ± 0.028 | 7.321 ± 0.03 |
1.437 ± 0.016 | 2.247 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
