
Oceanobacillus limi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Oceanobacillus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
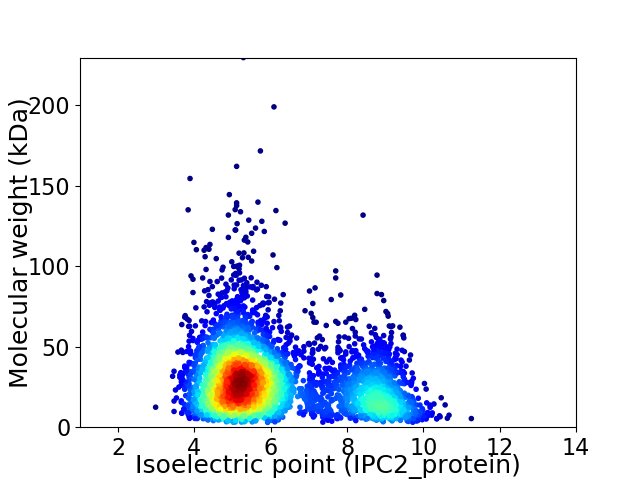
Virtual 2D-PAGE plot for 3982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0FAG6|A0A1I0FAG6_9BACI Catechol 2 3-dioxygenase OS=Oceanobacillus limi OX=930131 GN=SAMN05216389_11482 PE=4 SV=1
MM1 pKa = 7.5TKK3 pKa = 10.11TSITKK8 pKa = 8.94WLFALMLAMVLVLAACSDD26 pKa = 3.96DD27 pKa = 3.63TSSDD31 pKa = 3.5DD32 pKa = 5.31SSNDD36 pKa = 3.31DD37 pKa = 3.89PNTEE41 pKa = 3.62EE42 pKa = 4.91SSNNEE47 pKa = 3.5NDD49 pKa = 3.64GNDD52 pKa = 3.62AEE54 pKa = 4.38EE55 pKa = 4.53EE56 pKa = 4.23EE57 pKa = 4.32PAEE60 pKa = 4.8DD61 pKa = 4.7DD62 pKa = 3.69GLYY65 pKa = 10.83SIDD68 pKa = 4.61DD69 pKa = 3.81FSMDD73 pKa = 3.08KK74 pKa = 10.7TNAGEE79 pKa = 4.29AIEE82 pKa = 5.54GGTLNYY88 pKa = 10.69GLVSDD93 pKa = 4.4TVFEE97 pKa = 4.28GTLNFNFYY105 pKa = 10.66SGNPDD110 pKa = 2.94WEE112 pKa = 4.5VISWFDD118 pKa = 3.91EE119 pKa = 4.05PLLYY123 pKa = 10.38TDD125 pKa = 5.35ANYY128 pKa = 9.61TYY130 pKa = 9.78TNEE133 pKa = 4.05GPATYY138 pKa = 9.65EE139 pKa = 3.77VSEE142 pKa = 5.12DD143 pKa = 3.35GLTFTLTIKK152 pKa = 10.95DD153 pKa = 3.49GVNWHH158 pKa = 7.14DD159 pKa = 4.06GEE161 pKa = 4.53PVTAEE166 pKa = 3.6DD167 pKa = 2.88WAYY170 pKa = 8.61SYY172 pKa = 11.13EE173 pKa = 4.63VIGHH177 pKa = 6.93PDD179 pKa = 3.4YY180 pKa = 11.49TGVRR184 pKa = 11.84YY185 pKa = 10.14NDD187 pKa = 3.85DD188 pKa = 3.41FTNVVGMEE196 pKa = 4.15EE197 pKa = 4.25YY198 pKa = 10.45HH199 pKa = 7.73AGEE202 pKa = 4.32ADD204 pKa = 4.14EE205 pKa = 4.64ISGIEE210 pKa = 3.98IVDD213 pKa = 3.72EE214 pKa = 4.08KK215 pKa = 10.18TLKK218 pKa = 8.62ITHH221 pKa = 6.8HH222 pKa = 5.53EE223 pKa = 4.27TGPSMVTGGIWTYY236 pKa = 11.87AMPKK240 pKa = 9.8HH241 pKa = 6.24LFSEE245 pKa = 4.44IPVAEE250 pKa = 4.25MEE252 pKa = 4.19EE253 pKa = 4.25SDD255 pKa = 4.74LVRR258 pKa = 11.84EE259 pKa = 4.43NPIGMGPFKK268 pKa = 10.94VEE270 pKa = 4.23SIVPGEE276 pKa = 4.06SVVYY280 pKa = 10.43SKK282 pKa = 11.64NEE284 pKa = 4.04DD285 pKa = 3.39YY286 pKa = 11.05WNGEE290 pKa = 3.9PKK292 pKa = 10.34LDD294 pKa = 3.67EE295 pKa = 4.28VVLKK299 pKa = 10.31VVAPEE304 pKa = 4.02TVVNEE309 pKa = 4.22LEE311 pKa = 4.27TGGVDD316 pKa = 4.36LVNSFPTDD324 pKa = 3.5QFPDD328 pKa = 3.51NADD331 pKa = 3.44MSNVEE336 pKa = 4.3YY337 pKa = 11.02LGMIDD342 pKa = 4.35NAYY345 pKa = 9.41TYY347 pKa = 10.51IGFKK351 pKa = 10.12LGHH354 pKa = 6.73WDD356 pKa = 3.31ADD358 pKa = 3.52AGEE361 pKa = 4.47VVMDD365 pKa = 5.34PDD367 pKa = 4.8AKK369 pKa = 10.39MADD372 pKa = 3.53VNLRR376 pKa = 11.84RR377 pKa = 11.84AMWHH381 pKa = 6.36AVDD384 pKa = 3.66NDD386 pKa = 3.49AVGRR390 pKa = 11.84EE391 pKa = 4.44FYY393 pKa = 11.05NGLRR397 pKa = 11.84WNATSLIIPFFPDD410 pKa = 3.49YY411 pKa = 11.41YY412 pKa = 10.83DD413 pKa = 3.82DD414 pKa = 5.15TIDD417 pKa = 5.4APTFDD422 pKa = 4.39PDD424 pKa = 3.15EE425 pKa = 4.81ANRR428 pKa = 11.84LLDD431 pKa = 3.47EE432 pKa = 5.25AGYY435 pKa = 11.09EE436 pKa = 4.15DD437 pKa = 4.66VDD439 pKa = 3.75GDD441 pKa = 4.42GIRR444 pKa = 11.84EE445 pKa = 4.01NPEE448 pKa = 3.57GEE450 pKa = 3.87EE451 pKa = 3.85LVINFASMSGGDD463 pKa = 3.53TAEE466 pKa = 5.14PIANYY471 pKa = 9.89YY472 pKa = 8.09IQSWEE477 pKa = 3.93NVGLKK482 pKa = 10.25VEE484 pKa = 4.32LVDD487 pKa = 4.19GRR489 pKa = 11.84LMEE492 pKa = 4.65FNSFYY497 pKa = 11.34DD498 pKa = 3.27RR499 pKa = 11.84VEE501 pKa = 4.91ADD503 pKa = 4.8DD504 pKa = 4.81PDD506 pKa = 4.01IDD508 pKa = 4.01IYY510 pKa = 10.83QGAWGTGTDD519 pKa = 3.79VNPSGLYY526 pKa = 9.94GRR528 pKa = 11.84NASYY532 pKa = 10.93NYY534 pKa = 9.25PRR536 pKa = 11.84YY537 pKa = 10.01ASEE540 pKa = 4.02EE541 pKa = 3.88NDD543 pKa = 3.29RR544 pKa = 11.84LLAEE548 pKa = 4.88GNSEE552 pKa = 3.88AALDD556 pKa = 3.56VEE558 pKa = 4.73YY559 pKa = 10.64RR560 pKa = 11.84QEE562 pKa = 5.07VYY564 pKa = 10.78SEE566 pKa = 3.92WQQLMVEE573 pKa = 5.23DD574 pKa = 4.23IPVFPTLYY582 pKa = 9.07RR583 pKa = 11.84SVVIPVNEE591 pKa = 3.72RR592 pKa = 11.84VINYY596 pKa = 9.85SIAPDD601 pKa = 3.17KK602 pKa = 10.83YY603 pKa = 8.57IRR605 pKa = 11.84HH606 pKa = 6.66EE607 pKa = 3.79IAVTQEE613 pKa = 3.92EE614 pKa = 4.83PVVAEE619 pKa = 4.13
MM1 pKa = 7.5TKK3 pKa = 10.11TSITKK8 pKa = 8.94WLFALMLAMVLVLAACSDD26 pKa = 3.96DD27 pKa = 3.63TSSDD31 pKa = 3.5DD32 pKa = 5.31SSNDD36 pKa = 3.31DD37 pKa = 3.89PNTEE41 pKa = 3.62EE42 pKa = 4.91SSNNEE47 pKa = 3.5NDD49 pKa = 3.64GNDD52 pKa = 3.62AEE54 pKa = 4.38EE55 pKa = 4.53EE56 pKa = 4.23EE57 pKa = 4.32PAEE60 pKa = 4.8DD61 pKa = 4.7DD62 pKa = 3.69GLYY65 pKa = 10.83SIDD68 pKa = 4.61DD69 pKa = 3.81FSMDD73 pKa = 3.08KK74 pKa = 10.7TNAGEE79 pKa = 4.29AIEE82 pKa = 5.54GGTLNYY88 pKa = 10.69GLVSDD93 pKa = 4.4TVFEE97 pKa = 4.28GTLNFNFYY105 pKa = 10.66SGNPDD110 pKa = 2.94WEE112 pKa = 4.5VISWFDD118 pKa = 3.91EE119 pKa = 4.05PLLYY123 pKa = 10.38TDD125 pKa = 5.35ANYY128 pKa = 9.61TYY130 pKa = 9.78TNEE133 pKa = 4.05GPATYY138 pKa = 9.65EE139 pKa = 3.77VSEE142 pKa = 5.12DD143 pKa = 3.35GLTFTLTIKK152 pKa = 10.95DD153 pKa = 3.49GVNWHH158 pKa = 7.14DD159 pKa = 4.06GEE161 pKa = 4.53PVTAEE166 pKa = 3.6DD167 pKa = 2.88WAYY170 pKa = 8.61SYY172 pKa = 11.13EE173 pKa = 4.63VIGHH177 pKa = 6.93PDD179 pKa = 3.4YY180 pKa = 11.49TGVRR184 pKa = 11.84YY185 pKa = 10.14NDD187 pKa = 3.85DD188 pKa = 3.41FTNVVGMEE196 pKa = 4.15EE197 pKa = 4.25YY198 pKa = 10.45HH199 pKa = 7.73AGEE202 pKa = 4.32ADD204 pKa = 4.14EE205 pKa = 4.64ISGIEE210 pKa = 3.98IVDD213 pKa = 3.72EE214 pKa = 4.08KK215 pKa = 10.18TLKK218 pKa = 8.62ITHH221 pKa = 6.8HH222 pKa = 5.53EE223 pKa = 4.27TGPSMVTGGIWTYY236 pKa = 11.87AMPKK240 pKa = 9.8HH241 pKa = 6.24LFSEE245 pKa = 4.44IPVAEE250 pKa = 4.25MEE252 pKa = 4.19EE253 pKa = 4.25SDD255 pKa = 4.74LVRR258 pKa = 11.84EE259 pKa = 4.43NPIGMGPFKK268 pKa = 10.94VEE270 pKa = 4.23SIVPGEE276 pKa = 4.06SVVYY280 pKa = 10.43SKK282 pKa = 11.64NEE284 pKa = 4.04DD285 pKa = 3.39YY286 pKa = 11.05WNGEE290 pKa = 3.9PKK292 pKa = 10.34LDD294 pKa = 3.67EE295 pKa = 4.28VVLKK299 pKa = 10.31VVAPEE304 pKa = 4.02TVVNEE309 pKa = 4.22LEE311 pKa = 4.27TGGVDD316 pKa = 4.36LVNSFPTDD324 pKa = 3.5QFPDD328 pKa = 3.51NADD331 pKa = 3.44MSNVEE336 pKa = 4.3YY337 pKa = 11.02LGMIDD342 pKa = 4.35NAYY345 pKa = 9.41TYY347 pKa = 10.51IGFKK351 pKa = 10.12LGHH354 pKa = 6.73WDD356 pKa = 3.31ADD358 pKa = 3.52AGEE361 pKa = 4.47VVMDD365 pKa = 5.34PDD367 pKa = 4.8AKK369 pKa = 10.39MADD372 pKa = 3.53VNLRR376 pKa = 11.84RR377 pKa = 11.84AMWHH381 pKa = 6.36AVDD384 pKa = 3.66NDD386 pKa = 3.49AVGRR390 pKa = 11.84EE391 pKa = 4.44FYY393 pKa = 11.05NGLRR397 pKa = 11.84WNATSLIIPFFPDD410 pKa = 3.49YY411 pKa = 11.41YY412 pKa = 10.83DD413 pKa = 3.82DD414 pKa = 5.15TIDD417 pKa = 5.4APTFDD422 pKa = 4.39PDD424 pKa = 3.15EE425 pKa = 4.81ANRR428 pKa = 11.84LLDD431 pKa = 3.47EE432 pKa = 5.25AGYY435 pKa = 11.09EE436 pKa = 4.15DD437 pKa = 4.66VDD439 pKa = 3.75GDD441 pKa = 4.42GIRR444 pKa = 11.84EE445 pKa = 4.01NPEE448 pKa = 3.57GEE450 pKa = 3.87EE451 pKa = 3.85LVINFASMSGGDD463 pKa = 3.53TAEE466 pKa = 5.14PIANYY471 pKa = 9.89YY472 pKa = 8.09IQSWEE477 pKa = 3.93NVGLKK482 pKa = 10.25VEE484 pKa = 4.32LVDD487 pKa = 4.19GRR489 pKa = 11.84LMEE492 pKa = 4.65FNSFYY497 pKa = 11.34DD498 pKa = 3.27RR499 pKa = 11.84VEE501 pKa = 4.91ADD503 pKa = 4.8DD504 pKa = 4.81PDD506 pKa = 4.01IDD508 pKa = 4.01IYY510 pKa = 10.83QGAWGTGTDD519 pKa = 3.79VNPSGLYY526 pKa = 9.94GRR528 pKa = 11.84NASYY532 pKa = 10.93NYY534 pKa = 9.25PRR536 pKa = 11.84YY537 pKa = 10.01ASEE540 pKa = 4.02EE541 pKa = 3.88NDD543 pKa = 3.29RR544 pKa = 11.84LLAEE548 pKa = 4.88GNSEE552 pKa = 3.88AALDD556 pKa = 3.56VEE558 pKa = 4.73YY559 pKa = 10.64RR560 pKa = 11.84QEE562 pKa = 5.07VYY564 pKa = 10.78SEE566 pKa = 3.92WQQLMVEE573 pKa = 5.23DD574 pKa = 4.23IPVFPTLYY582 pKa = 9.07RR583 pKa = 11.84SVVIPVNEE591 pKa = 3.72RR592 pKa = 11.84VINYY596 pKa = 9.85SIAPDD601 pKa = 3.17KK602 pKa = 10.83YY603 pKa = 8.57IRR605 pKa = 11.84HH606 pKa = 6.66EE607 pKa = 3.79IAVTQEE613 pKa = 3.92EE614 pKa = 4.83PVVAEE619 pKa = 4.13
Molecular weight: 69.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0GWR6|A0A1I0GWR6_9BACI Non-homologous end joining protein Ku OS=Oceanobacillus limi OX=930131 GN=ku PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 7.99KK14 pKa = 9.24VHH16 pKa = 5.91GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTKK25 pKa = 10.15NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 7.99KK14 pKa = 9.24VHH16 pKa = 5.91GFRR19 pKa = 11.84QRR21 pKa = 11.84MSTKK25 pKa = 10.15NGRR28 pKa = 11.84KK29 pKa = 8.49VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1095241 |
27 |
1965 |
275.0 |
31.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.4 ± 0.043 | 0.618 ± 0.011 |
5.541 ± 0.035 | 7.852 ± 0.043 |
4.587 ± 0.038 | 6.63 ± 0.035 |
2.105 ± 0.019 | 8.119 ± 0.039 |
6.696 ± 0.042 | 9.489 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.779 ± 0.018 | 4.781 ± 0.029 |
3.44 ± 0.024 | 3.812 ± 0.03 |
3.96 ± 0.032 | 6.021 ± 0.026 |
5.483 ± 0.028 | 6.965 ± 0.032 |
1.031 ± 0.016 | 3.692 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
