
Gracilibacillus dipsosauri
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli;
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
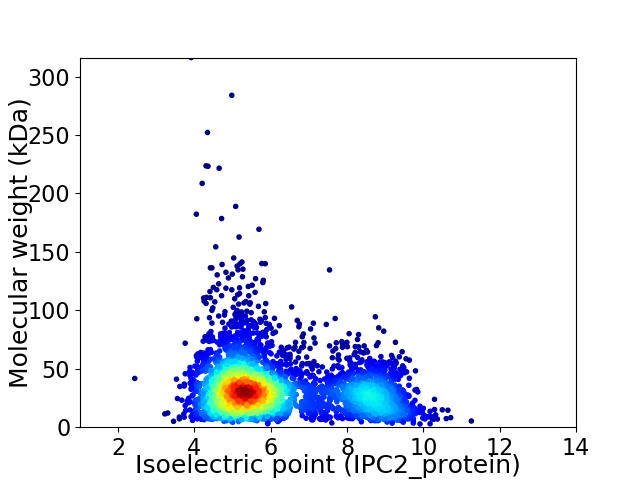
Virtual 2D-PAGE plot for 3941 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317L148|A0A317L148_9BACI Aminotransferase class V-fold PLP-dependent enzyme OS=Gracilibacillus dipsosauri OX=178340 GN=DLJ74_11320 PE=3 SV=1
MM1 pKa = 7.32SKK3 pKa = 9.7WKK5 pKa = 10.85LMMLVAVTLLTVGLLAACGNAPDD28 pKa = 5.45DD29 pKa = 5.05AGSDD33 pKa = 3.79EE34 pKa = 4.48NNEE37 pKa = 4.0ANASEE42 pKa = 4.17NGEE45 pKa = 4.02GSGDD49 pKa = 3.53EE50 pKa = 4.06GSEE53 pKa = 4.18DD54 pKa = 4.21GEE56 pKa = 4.23VSGPVNIDD64 pKa = 2.9GSSTVLPIMEE74 pKa = 4.51YY75 pKa = 9.38LTYY78 pKa = 10.22EE79 pKa = 4.21YY80 pKa = 10.62NQEE83 pKa = 3.99NPEE86 pKa = 4.03VEE88 pKa = 4.18TALNSSGSGGGFEE101 pKa = 4.38KK102 pKa = 10.19STVGEE107 pKa = 3.98IDD109 pKa = 3.68LSNASRR115 pKa = 11.84PIKK118 pKa = 10.55DD119 pKa = 3.52EE120 pKa = 4.05EE121 pKa = 4.32KK122 pKa = 10.42ATAEE126 pKa = 4.0EE127 pKa = 4.1NGITLHH133 pKa = 6.35EE134 pKa = 4.76IEE136 pKa = 4.92LAYY139 pKa = 10.69DD140 pKa = 3.66GLSVVVSQEE149 pKa = 3.37NDD151 pKa = 3.14FVEE154 pKa = 4.61NLTIDD159 pKa = 3.36QLKK162 pKa = 10.59QIFLADD168 pKa = 3.72GGATKK173 pKa = 10.22WSDD176 pKa = 3.66INPEE180 pKa = 3.84WPEE183 pKa = 3.81EE184 pKa = 4.35DD185 pKa = 2.96IVIYY189 pKa = 10.82SPGHH193 pKa = 6.25DD194 pKa = 4.07SGTFDD199 pKa = 3.78YY200 pKa = 10.64FNEE203 pKa = 4.53VILEE207 pKa = 4.1EE208 pKa = 4.18NPMRR212 pKa = 11.84EE213 pKa = 4.33GEE215 pKa = 4.21NVTLSEE221 pKa = 5.11DD222 pKa = 3.95DD223 pKa = 3.59NTLVTGIQSDD233 pKa = 4.93PYY235 pKa = 9.5AIGYY239 pKa = 8.87FGYY242 pKa = 10.6AYY244 pKa = 10.42YY245 pKa = 10.99AEE247 pKa = 4.46NQDD250 pKa = 3.67TLKK253 pKa = 11.13VLGIDD258 pKa = 3.79NGDD261 pKa = 3.75GNPVKK266 pKa = 9.6PTGDD270 pKa = 3.82TIQDD274 pKa = 3.31GSYY277 pKa = 10.78SPLSRR282 pKa = 11.84PLFTYY287 pKa = 10.86VNVDD291 pKa = 3.29YY292 pKa = 9.99MKK294 pKa = 10.58EE295 pKa = 3.96KK296 pKa = 10.32PQVLDD301 pKa = 3.82FVKK304 pKa = 10.65FSIEE308 pKa = 3.9NAGAAAEE315 pKa = 4.16EE316 pKa = 4.37VGYY319 pKa = 10.72VALPDD324 pKa = 3.56EE325 pKa = 5.32KK326 pKa = 11.02YY327 pKa = 10.01QEE329 pKa = 3.91QLTEE333 pKa = 4.01IEE335 pKa = 4.19EE336 pKa = 4.25LAAEE340 pKa = 4.21
MM1 pKa = 7.32SKK3 pKa = 9.7WKK5 pKa = 10.85LMMLVAVTLLTVGLLAACGNAPDD28 pKa = 5.45DD29 pKa = 5.05AGSDD33 pKa = 3.79EE34 pKa = 4.48NNEE37 pKa = 4.0ANASEE42 pKa = 4.17NGEE45 pKa = 4.02GSGDD49 pKa = 3.53EE50 pKa = 4.06GSEE53 pKa = 4.18DD54 pKa = 4.21GEE56 pKa = 4.23VSGPVNIDD64 pKa = 2.9GSSTVLPIMEE74 pKa = 4.51YY75 pKa = 9.38LTYY78 pKa = 10.22EE79 pKa = 4.21YY80 pKa = 10.62NQEE83 pKa = 3.99NPEE86 pKa = 4.03VEE88 pKa = 4.18TALNSSGSGGGFEE101 pKa = 4.38KK102 pKa = 10.19STVGEE107 pKa = 3.98IDD109 pKa = 3.68LSNASRR115 pKa = 11.84PIKK118 pKa = 10.55DD119 pKa = 3.52EE120 pKa = 4.05EE121 pKa = 4.32KK122 pKa = 10.42ATAEE126 pKa = 4.0EE127 pKa = 4.1NGITLHH133 pKa = 6.35EE134 pKa = 4.76IEE136 pKa = 4.92LAYY139 pKa = 10.69DD140 pKa = 3.66GLSVVVSQEE149 pKa = 3.37NDD151 pKa = 3.14FVEE154 pKa = 4.61NLTIDD159 pKa = 3.36QLKK162 pKa = 10.59QIFLADD168 pKa = 3.72GGATKK173 pKa = 10.22WSDD176 pKa = 3.66INPEE180 pKa = 3.84WPEE183 pKa = 3.81EE184 pKa = 4.35DD185 pKa = 2.96IVIYY189 pKa = 10.82SPGHH193 pKa = 6.25DD194 pKa = 4.07SGTFDD199 pKa = 3.78YY200 pKa = 10.64FNEE203 pKa = 4.53VILEE207 pKa = 4.1EE208 pKa = 4.18NPMRR212 pKa = 11.84EE213 pKa = 4.33GEE215 pKa = 4.21NVTLSEE221 pKa = 5.11DD222 pKa = 3.95DD223 pKa = 3.59NTLVTGIQSDD233 pKa = 4.93PYY235 pKa = 9.5AIGYY239 pKa = 8.87FGYY242 pKa = 10.6AYY244 pKa = 10.42YY245 pKa = 10.99AEE247 pKa = 4.46NQDD250 pKa = 3.67TLKK253 pKa = 11.13VLGIDD258 pKa = 3.79NGDD261 pKa = 3.75GNPVKK266 pKa = 9.6PTGDD270 pKa = 3.82TIQDD274 pKa = 3.31GSYY277 pKa = 10.78SPLSRR282 pKa = 11.84PLFTYY287 pKa = 10.86VNVDD291 pKa = 3.29YY292 pKa = 9.99MKK294 pKa = 10.58EE295 pKa = 3.96KK296 pKa = 10.32PQVLDD301 pKa = 3.82FVKK304 pKa = 10.65FSIEE308 pKa = 3.9NAGAAAEE315 pKa = 4.16EE316 pKa = 4.37VGYY319 pKa = 10.72VALPDD324 pKa = 3.56EE325 pKa = 5.32KK326 pKa = 11.02YY327 pKa = 10.01QEE329 pKa = 3.91QLTEE333 pKa = 4.01IEE335 pKa = 4.19EE336 pKa = 4.25LAAEE340 pKa = 4.21
Molecular weight: 36.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317L4B9|A0A317L4B9_9BACI DUF2935 domain-containing protein OS=Gracilibacillus dipsosauri OX=178340 GN=DLJ74_00500 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.66NGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.38KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 9.66NGRR28 pKa = 11.84RR29 pKa = 11.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.38KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1208377 |
24 |
2921 |
306.6 |
34.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.513 ± 0.043 | 0.657 ± 0.011 |
5.347 ± 0.032 | 7.636 ± 0.044 |
4.603 ± 0.032 | 6.484 ± 0.036 |
2.185 ± 0.018 | 8.276 ± 0.043 |
6.808 ± 0.033 | 9.726 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.752 ± 0.018 | 4.667 ± 0.03 |
3.544 ± 0.021 | 4.087 ± 0.031 |
3.802 ± 0.028 | 5.941 ± 0.028 |
5.435 ± 0.03 | 6.605 ± 0.03 |
1.188 ± 0.017 | 3.747 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
