
Torque teno virus 24
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
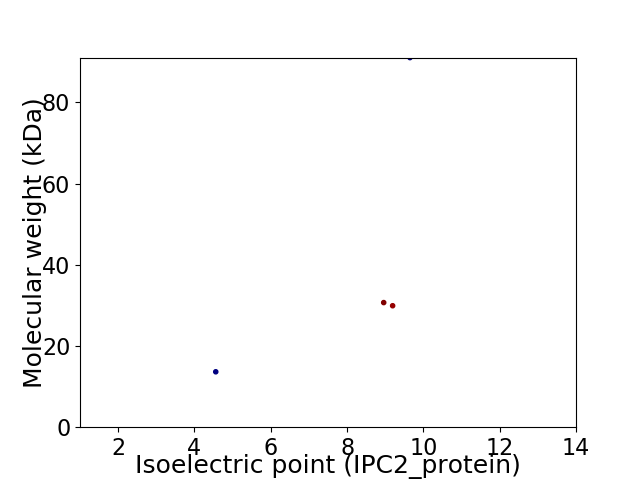
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
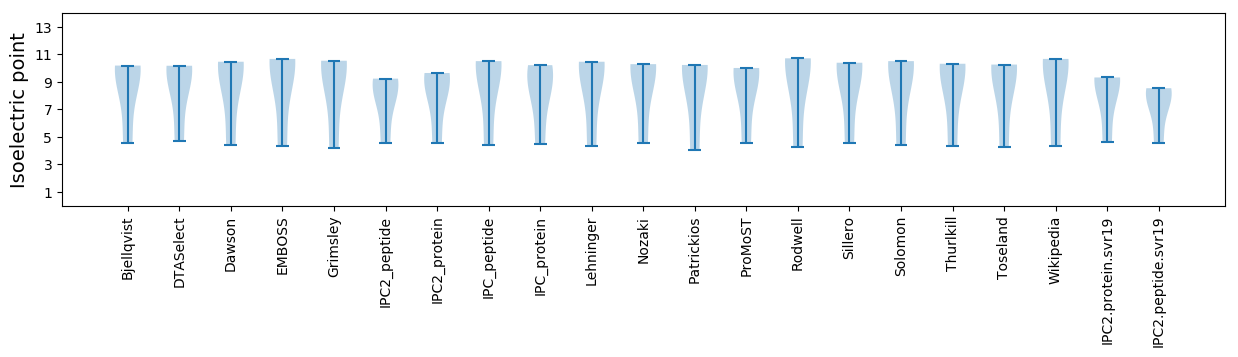
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91CY7|Q91CY7_9VIRU ORF4 ORF3 ORF2 ORF1 genes clone: SAa-01 OS=Torque teno virus 24 OX=687363 PE=4 SV=1
MM1 pKa = 7.37SIWRR5 pKa = 11.84PPLGNVSFRR14 pKa = 11.84EE15 pKa = 4.19RR16 pKa = 11.84NWLQACDD23 pKa = 3.54QSHH26 pKa = 6.04ATFCGCGDD34 pKa = 5.13FILHH38 pKa = 6.11LTNLAARR45 pKa = 11.84FALQGPPPPGGPPRR59 pKa = 11.84PRR61 pKa = 11.84PPLLRR66 pKa = 11.84ALPAPEE72 pKa = 3.78VRR74 pKa = 11.84RR75 pKa = 11.84EE76 pKa = 3.85TRR78 pKa = 11.84TEE80 pKa = 3.68NPGASDD86 pKa = 4.18EE87 pKa = 4.29PWPGDD92 pKa = 3.43GGGSDD97 pKa = 3.36AAAGGRR103 pKa = 11.84AAGDD107 pKa = 3.35GGDD110 pKa = 4.44AYY112 pKa = 10.9DD113 pKa = 5.38AGDD116 pKa = 4.83LEE118 pKa = 5.03DD119 pKa = 5.46LFAAVDD125 pKa = 3.77EE126 pKa = 4.62EE127 pKa = 4.46QQQ129 pKa = 3.22
MM1 pKa = 7.37SIWRR5 pKa = 11.84PPLGNVSFRR14 pKa = 11.84EE15 pKa = 4.19RR16 pKa = 11.84NWLQACDD23 pKa = 3.54QSHH26 pKa = 6.04ATFCGCGDD34 pKa = 5.13FILHH38 pKa = 6.11LTNLAARR45 pKa = 11.84FALQGPPPPGGPPRR59 pKa = 11.84PRR61 pKa = 11.84PPLLRR66 pKa = 11.84ALPAPEE72 pKa = 3.78VRR74 pKa = 11.84RR75 pKa = 11.84EE76 pKa = 3.85TRR78 pKa = 11.84TEE80 pKa = 3.68NPGASDD86 pKa = 4.18EE87 pKa = 4.29PWPGDD92 pKa = 3.43GGGSDD97 pKa = 3.36AAAGGRR103 pKa = 11.84AAGDD107 pKa = 3.35GGDD110 pKa = 4.44AYY112 pKa = 10.9DD113 pKa = 5.38AGDD116 pKa = 4.83LEE118 pKa = 5.03DD119 pKa = 5.46LFAAVDD125 pKa = 3.77EE126 pKa = 4.62EE127 pKa = 4.46QQQ129 pKa = 3.22
Molecular weight: 13.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91CY6|Q91CY6_9VIRU ORF4 ORF3 ORF2 ORF1 genes clone: SAa-01 OS=Torque teno virus 24 OX=687363 PE=4 SV=1
MM1 pKa = 7.54AWRR4 pKa = 11.84WWWQRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WWPRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84WRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84PVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84ATVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84WRR48 pKa = 11.84GRR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84RR54 pKa = 11.84TYY56 pKa = 8.33TRR58 pKa = 11.84RR59 pKa = 11.84AVRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84PKK68 pKa = 10.02KK69 pKa = 10.43KK70 pKa = 10.23LVLTQWSPQTVRR82 pKa = 11.84NCTIRR87 pKa = 11.84GIVPMVICGHH97 pKa = 5.32TRR99 pKa = 11.84AGRR102 pKa = 11.84NFAIHH107 pKa = 6.6SEE109 pKa = 4.22DD110 pKa = 3.29FTSQIKK116 pKa = 9.04PHH118 pKa = 6.53GGSFSTTTWSLKK130 pKa = 9.29VCWDD134 pKa = 3.37EE135 pKa = 4.02HH136 pKa = 5.33QKK138 pKa = 10.7FMNRR142 pKa = 11.84WSYY145 pKa = 11.68PNTQLDD151 pKa = 3.68LARR154 pKa = 11.84YY155 pKa = 9.4LGVTFYY161 pKa = 10.48FYY163 pKa = 10.8RR164 pKa = 11.84DD165 pKa = 3.15QKK167 pKa = 9.77TDD169 pKa = 4.12YY170 pKa = 10.19IVQWSRR176 pKa = 11.84NPPFKK181 pKa = 10.62LNKK184 pKa = 9.25YY185 pKa = 8.73SSPMYY190 pKa = 9.82HH191 pKa = 7.45PGMMMQARR199 pKa = 11.84KK200 pKa = 10.27KK201 pKa = 10.87LMVPSFQTRR210 pKa = 11.84PRR212 pKa = 11.84GRR214 pKa = 11.84KK215 pKa = 8.5RR216 pKa = 11.84YY217 pKa = 8.94RR218 pKa = 11.84VRR220 pKa = 11.84IRR222 pKa = 11.84PPNMFADD229 pKa = 3.28KK230 pKa = 10.2WYY232 pKa = 8.44TQEE235 pKa = 4.64DD236 pKa = 4.86LCPVPLVQLVVSAASFMHH254 pKa = 6.59PFCSPLTNNPCITFQVLKK272 pKa = 10.53DD273 pKa = 3.7NYY275 pKa = 9.29YY276 pKa = 10.49SCIGVNSLEE285 pKa = 4.23THH287 pKa = 6.0KK288 pKa = 11.14NSYY291 pKa = 9.09TKK293 pKa = 10.56LQEE296 pKa = 3.82WLYY299 pKa = 9.29KK300 pKa = 10.26TNTFFEE306 pKa = 4.64TTQVIAQLSPSFAPAYY322 pKa = 9.69KK323 pKa = 10.54PNGTNNNTHH332 pKa = 6.46KK333 pKa = 10.12PLGGMVPEE341 pKa = 4.78LKK343 pKa = 10.66YY344 pKa = 11.32NNAGFHH350 pKa = 6.1TGNNPVFGMCKK361 pKa = 9.85YY362 pKa = 10.45KK363 pKa = 10.68PSTLTMQNANKK374 pKa = 9.26WFWEE378 pKa = 3.89NLQVEE383 pKa = 4.65NDD385 pKa = 3.65LQSKK389 pKa = 9.0YY390 pKa = 10.94GKK392 pKa = 10.67ANLTCMEE399 pKa = 4.51YY400 pKa = 8.3HH401 pKa = 5.95TGIYY405 pKa = 10.2SSIFLSPQRR414 pKa = 11.84NLQFPAAYY422 pKa = 10.46LDD424 pKa = 3.5VTYY427 pKa = 11.1NPNCDD432 pKa = 2.98RR433 pKa = 11.84GIGNKK438 pKa = 8.53IWFQYY443 pKa = 6.58STKK446 pKa = 8.33MTTDD450 pKa = 4.47FEE452 pKa = 4.5QKK454 pKa = 9.03QCKK457 pKa = 9.15CVLEE461 pKa = 4.85NIPLWSAFHH470 pKa = 7.03GYY472 pKa = 9.75PDD474 pKa = 5.47FIEE477 pKa = 4.31QEE479 pKa = 4.09LSISAEE485 pKa = 3.51IHH487 pKa = 6.03NFGLVCFICPYY498 pKa = 9.64TFPPCYY504 pKa = 10.25NKK506 pKa = 9.82TKK508 pKa = 10.27PEE510 pKa = 3.86QGYY513 pKa = 8.55VFYY516 pKa = 9.41DD517 pKa = 3.43TTFGNGKK524 pKa = 8.52MPDD527 pKa = 3.68GSGHH531 pKa = 6.5IPIYY535 pKa = 8.8WQQRR539 pKa = 11.84WFIRR543 pKa = 11.84LAFQTQVMHH552 pKa = 6.94DD553 pKa = 4.07LVMCGPFSYY562 pKa = 10.9KK563 pKa = 10.35DD564 pKa = 3.48DD565 pKa = 4.36LANTTITARR574 pKa = 11.84YY575 pKa = 8.8KK576 pKa = 10.79FRR578 pKa = 11.84FKK580 pKa = 10.21WGGNIIPEE588 pKa = 4.09QIIKK592 pKa = 10.16NPCHH596 pKa = 7.07RR597 pKa = 11.84EE598 pKa = 3.67QSLASYY604 pKa = 10.19PDD606 pKa = 3.17RR607 pKa = 11.84QRR609 pKa = 11.84RR610 pKa = 11.84DD611 pKa = 3.57LQVVDD616 pKa = 4.78PSTMGPIYY624 pKa = 10.06TFHH627 pKa = 5.7TWDD630 pKa = 2.87WRR632 pKa = 11.84RR633 pKa = 11.84GLFGTDD639 pKa = 3.43AIQRR643 pKa = 11.84VSQKK647 pKa = 10.43PGDD650 pKa = 3.89ALRR653 pKa = 11.84ITNPFKK659 pKa = 10.62RR660 pKa = 11.84PRR662 pKa = 11.84YY663 pKa = 9.1VPPTDD668 pKa = 3.52RR669 pKa = 11.84EE670 pKa = 4.27DD671 pKa = 3.45YY672 pKa = 10.59RR673 pKa = 11.84QEE675 pKa = 3.6EE676 pKa = 4.36DD677 pKa = 3.59FALQEE682 pKa = 3.92KK683 pKa = 10.1RR684 pKa = 11.84RR685 pKa = 11.84RR686 pKa = 11.84TSTEE690 pKa = 3.39EE691 pKa = 3.98AQDD694 pKa = 3.83EE695 pKa = 4.32EE696 pKa = 5.23SPPQSAPLLQQRR708 pKa = 11.84QQRR711 pKa = 11.84EE712 pKa = 4.04LSVQLAEE719 pKa = 4.19QQRR722 pKa = 11.84LGHH725 pKa = 5.59QLRR728 pKa = 11.84FILQEE733 pKa = 3.91VLKK736 pKa = 8.68TQAGLHH742 pKa = 6.02LNPLLLGPRR751 pKa = 11.84PTRR754 pKa = 11.84SISLSPLEE762 pKa = 4.86DD763 pKa = 3.32SSPP766 pKa = 3.34
MM1 pKa = 7.54AWRR4 pKa = 11.84WWWQRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84WWPRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84WRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84PVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84ATVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84WRR48 pKa = 11.84GRR50 pKa = 11.84RR51 pKa = 11.84GRR53 pKa = 11.84RR54 pKa = 11.84TYY56 pKa = 8.33TRR58 pKa = 11.84RR59 pKa = 11.84AVRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84RR66 pKa = 11.84PKK68 pKa = 10.02KK69 pKa = 10.43KK70 pKa = 10.23LVLTQWSPQTVRR82 pKa = 11.84NCTIRR87 pKa = 11.84GIVPMVICGHH97 pKa = 5.32TRR99 pKa = 11.84AGRR102 pKa = 11.84NFAIHH107 pKa = 6.6SEE109 pKa = 4.22DD110 pKa = 3.29FTSQIKK116 pKa = 9.04PHH118 pKa = 6.53GGSFSTTTWSLKK130 pKa = 9.29VCWDD134 pKa = 3.37EE135 pKa = 4.02HH136 pKa = 5.33QKK138 pKa = 10.7FMNRR142 pKa = 11.84WSYY145 pKa = 11.68PNTQLDD151 pKa = 3.68LARR154 pKa = 11.84YY155 pKa = 9.4LGVTFYY161 pKa = 10.48FYY163 pKa = 10.8RR164 pKa = 11.84DD165 pKa = 3.15QKK167 pKa = 9.77TDD169 pKa = 4.12YY170 pKa = 10.19IVQWSRR176 pKa = 11.84NPPFKK181 pKa = 10.62LNKK184 pKa = 9.25YY185 pKa = 8.73SSPMYY190 pKa = 9.82HH191 pKa = 7.45PGMMMQARR199 pKa = 11.84KK200 pKa = 10.27KK201 pKa = 10.87LMVPSFQTRR210 pKa = 11.84PRR212 pKa = 11.84GRR214 pKa = 11.84KK215 pKa = 8.5RR216 pKa = 11.84YY217 pKa = 8.94RR218 pKa = 11.84VRR220 pKa = 11.84IRR222 pKa = 11.84PPNMFADD229 pKa = 3.28KK230 pKa = 10.2WYY232 pKa = 8.44TQEE235 pKa = 4.64DD236 pKa = 4.86LCPVPLVQLVVSAASFMHH254 pKa = 6.59PFCSPLTNNPCITFQVLKK272 pKa = 10.53DD273 pKa = 3.7NYY275 pKa = 9.29YY276 pKa = 10.49SCIGVNSLEE285 pKa = 4.23THH287 pKa = 6.0KK288 pKa = 11.14NSYY291 pKa = 9.09TKK293 pKa = 10.56LQEE296 pKa = 3.82WLYY299 pKa = 9.29KK300 pKa = 10.26TNTFFEE306 pKa = 4.64TTQVIAQLSPSFAPAYY322 pKa = 9.69KK323 pKa = 10.54PNGTNNNTHH332 pKa = 6.46KK333 pKa = 10.12PLGGMVPEE341 pKa = 4.78LKK343 pKa = 10.66YY344 pKa = 11.32NNAGFHH350 pKa = 6.1TGNNPVFGMCKK361 pKa = 9.85YY362 pKa = 10.45KK363 pKa = 10.68PSTLTMQNANKK374 pKa = 9.26WFWEE378 pKa = 3.89NLQVEE383 pKa = 4.65NDD385 pKa = 3.65LQSKK389 pKa = 9.0YY390 pKa = 10.94GKK392 pKa = 10.67ANLTCMEE399 pKa = 4.51YY400 pKa = 8.3HH401 pKa = 5.95TGIYY405 pKa = 10.2SSIFLSPQRR414 pKa = 11.84NLQFPAAYY422 pKa = 10.46LDD424 pKa = 3.5VTYY427 pKa = 11.1NPNCDD432 pKa = 2.98RR433 pKa = 11.84GIGNKK438 pKa = 8.53IWFQYY443 pKa = 6.58STKK446 pKa = 8.33MTTDD450 pKa = 4.47FEE452 pKa = 4.5QKK454 pKa = 9.03QCKK457 pKa = 9.15CVLEE461 pKa = 4.85NIPLWSAFHH470 pKa = 7.03GYY472 pKa = 9.75PDD474 pKa = 5.47FIEE477 pKa = 4.31QEE479 pKa = 4.09LSISAEE485 pKa = 3.51IHH487 pKa = 6.03NFGLVCFICPYY498 pKa = 9.64TFPPCYY504 pKa = 10.25NKK506 pKa = 9.82TKK508 pKa = 10.27PEE510 pKa = 3.86QGYY513 pKa = 8.55VFYY516 pKa = 9.41DD517 pKa = 3.43TTFGNGKK524 pKa = 8.52MPDD527 pKa = 3.68GSGHH531 pKa = 6.5IPIYY535 pKa = 8.8WQQRR539 pKa = 11.84WFIRR543 pKa = 11.84LAFQTQVMHH552 pKa = 6.94DD553 pKa = 4.07LVMCGPFSYY562 pKa = 10.9KK563 pKa = 10.35DD564 pKa = 3.48DD565 pKa = 4.36LANTTITARR574 pKa = 11.84YY575 pKa = 8.8KK576 pKa = 10.79FRR578 pKa = 11.84FKK580 pKa = 10.21WGGNIIPEE588 pKa = 4.09QIIKK592 pKa = 10.16NPCHH596 pKa = 7.07RR597 pKa = 11.84EE598 pKa = 3.67QSLASYY604 pKa = 10.19PDD606 pKa = 3.17RR607 pKa = 11.84QRR609 pKa = 11.84RR610 pKa = 11.84DD611 pKa = 3.57LQVVDD616 pKa = 4.78PSTMGPIYY624 pKa = 10.06TFHH627 pKa = 5.7TWDD630 pKa = 2.87WRR632 pKa = 11.84RR633 pKa = 11.84GLFGTDD639 pKa = 3.43AIQRR643 pKa = 11.84VSQKK647 pKa = 10.43PGDD650 pKa = 3.89ALRR653 pKa = 11.84ITNPFKK659 pKa = 10.62RR660 pKa = 11.84PRR662 pKa = 11.84YY663 pKa = 9.1VPPTDD668 pKa = 3.52RR669 pKa = 11.84EE670 pKa = 4.27DD671 pKa = 3.45YY672 pKa = 10.59RR673 pKa = 11.84QEE675 pKa = 3.6EE676 pKa = 4.36DD677 pKa = 3.59FALQEE682 pKa = 3.92KK683 pKa = 10.1RR684 pKa = 11.84RR685 pKa = 11.84RR686 pKa = 11.84TSTEE690 pKa = 3.39EE691 pKa = 3.98AQDD694 pKa = 3.83EE695 pKa = 4.32EE696 pKa = 5.23SPPQSAPLLQQRR708 pKa = 11.84QQRR711 pKa = 11.84EE712 pKa = 4.04LSVQLAEE719 pKa = 4.19QQRR722 pKa = 11.84LGHH725 pKa = 5.59QLRR728 pKa = 11.84FILQEE733 pKa = 3.91VLKK736 pKa = 8.68TQAGLHH742 pKa = 6.02LNPLLLGPRR751 pKa = 11.84PTRR754 pKa = 11.84SISLSPLEE762 pKa = 4.86DD763 pKa = 3.32SSPP766 pKa = 3.34
Molecular weight: 91.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1452 |
129 |
766 |
363.0 |
41.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.231 ± 2.287 | 1.928 ± 0.233 |
5.028 ± 0.827 | 4.408 ± 0.354 |
4.201 ± 0.604 | 7.231 ± 1.543 |
2.135 ± 0.146 | 2.961 ± 0.786 |
4.408 ± 0.754 | 7.231 ± 0.198 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.791 ± 0.482 | 3.994 ± 0.722 |
9.711 ± 1.787 | 5.096 ± 1.034 |
10.468 ± 0.307 | 6.543 ± 1.519 |
6.474 ± 0.524 | 3.512 ± 0.605 |
2.548 ± 0.396 | 3.099 ± 0.949 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
