
Ajellomyces capsulatus (strain G186AR / H82 / ATCC MYA-2454 / RMSCC 2432) (Darling s disease fungus) (Histoplasma capsulatum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Onygenales; Ajellomycetaceae; Histoplasma; Histoplasma capsulatum
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
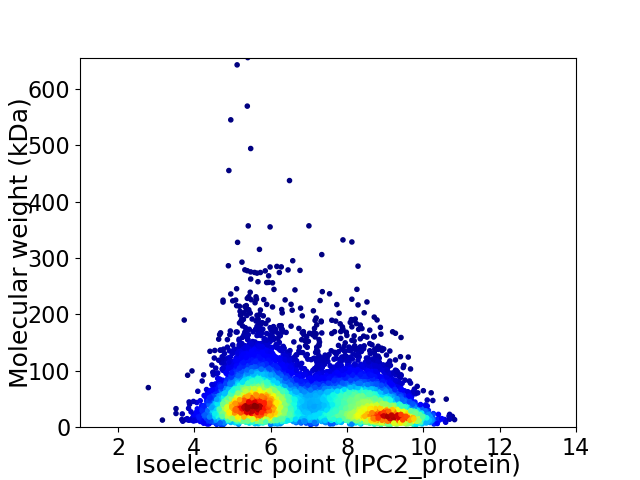
Virtual 2D-PAGE plot for 9214 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0NLH3|C0NLH3_AJECG MFS transporter OS=Ajellomyces capsulatus (strain G186AR / H82 / ATCC MYA-2454 / RMSCC 2432) OX=447093 GN=HCBG_04353 PE=4 SV=1
MM1 pKa = 6.53VTYY4 pKa = 10.51ILYY7 pKa = 9.99GFRR10 pKa = 11.84WNRR13 pKa = 11.84AANPLAPGIRR23 pKa = 11.84AYY25 pKa = 8.82ITLCNILDD33 pKa = 3.83AAAEE37 pKa = 4.16YY38 pKa = 9.9LQHH41 pKa = 7.2PSTTTAVLNSFKK53 pKa = 10.99LIDD56 pKa = 4.09SNILTHH62 pKa = 7.2LPDD65 pKa = 4.53LEE67 pKa = 5.99LIEE70 pKa = 5.0QYY72 pKa = 11.14DD73 pKa = 4.17PEE75 pKa = 5.37DD76 pKa = 3.77LSADD80 pKa = 3.56AVSQPYY86 pKa = 10.3AYY88 pKa = 10.29VAAKK92 pKa = 9.27TMTMGANALSGAGLGLSLQDD112 pKa = 5.16ILQQDD117 pKa = 4.11PGLSTAGTDD126 pKa = 3.42VFKK129 pKa = 11.11KK130 pKa = 10.59LRR132 pKa = 11.84DD133 pKa = 3.5EE134 pKa = 4.59LAPDD138 pKa = 3.95SEE140 pKa = 4.57IGWFVVYY147 pKa = 10.82NGDD150 pKa = 3.53PEE152 pKa = 4.06RR153 pKa = 11.84SYY155 pKa = 11.92GSFYY159 pKa = 10.85GDD161 pKa = 3.2SAVEE165 pKa = 3.99SDD167 pKa = 3.55GG168 pKa = 4.2
MM1 pKa = 6.53VTYY4 pKa = 10.51ILYY7 pKa = 9.99GFRR10 pKa = 11.84WNRR13 pKa = 11.84AANPLAPGIRR23 pKa = 11.84AYY25 pKa = 8.82ITLCNILDD33 pKa = 3.83AAAEE37 pKa = 4.16YY38 pKa = 9.9LQHH41 pKa = 7.2PSTTTAVLNSFKK53 pKa = 10.99LIDD56 pKa = 4.09SNILTHH62 pKa = 7.2LPDD65 pKa = 4.53LEE67 pKa = 5.99LIEE70 pKa = 5.0QYY72 pKa = 11.14DD73 pKa = 4.17PEE75 pKa = 5.37DD76 pKa = 3.77LSADD80 pKa = 3.56AVSQPYY86 pKa = 10.3AYY88 pKa = 10.29VAAKK92 pKa = 9.27TMTMGANALSGAGLGLSLQDD112 pKa = 5.16ILQQDD117 pKa = 4.11PGLSTAGTDD126 pKa = 3.42VFKK129 pKa = 11.11KK130 pKa = 10.59LRR132 pKa = 11.84DD133 pKa = 3.5EE134 pKa = 4.59LAPDD138 pKa = 3.95SEE140 pKa = 4.57IGWFVVYY147 pKa = 10.82NGDD150 pKa = 3.53PEE152 pKa = 4.06RR153 pKa = 11.84SYY155 pKa = 11.92GSFYY159 pKa = 10.85GDD161 pKa = 3.2SAVEE165 pKa = 3.99SDD167 pKa = 3.55GG168 pKa = 4.2
Molecular weight: 18.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0NPA9|C0NPA9_AJECG Uncharacterized protein OS=Ajellomyces capsulatus (strain G186AR / H82 / ATCC MYA-2454 / RMSCC 2432) OX=447093 GN=HCBG_04989 PE=4 SV=1
MM1 pKa = 7.29IAGFFFICWRR11 pKa = 11.84IGQLVTLIIPIGILSWFVDD30 pKa = 3.63GFVKK34 pKa = 10.62NNQLTPTYY42 pKa = 9.91ILVLFIVSVLGIFWALDD59 pKa = 3.12TLIRR63 pKa = 11.84HH64 pKa = 5.9SNAKK68 pKa = 9.99RR69 pKa = 11.84SAHH72 pKa = 5.27FVAFVDD78 pKa = 3.95LCFVGSFIAGVYY90 pKa = 6.8QLRR93 pKa = 11.84RR94 pKa = 11.84IANADD99 pKa = 3.37CGNFRR104 pKa = 11.84FDD106 pKa = 4.89PLTLSLGPFGFAGQSANNPLARR128 pKa = 11.84DD129 pKa = 3.84PTKK132 pKa = 10.66VCAMLKK138 pKa = 9.33TSFAFGIMNIGSFFITSILAVLMHH162 pKa = 5.97HH163 pKa = 6.36HH164 pKa = 6.59EE165 pKa = 4.29EE166 pKa = 4.32RR167 pKa = 11.84EE168 pKa = 4.17HH169 pKa = 6.45EE170 pKa = 4.41KK171 pKa = 10.69GSSRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84GSHH180 pKa = 5.08SSHH183 pKa = 6.83RR184 pKa = 11.84GHH186 pKa = 6.32SGSRR190 pKa = 11.84HH191 pKa = 4.98RR192 pKa = 11.84RR193 pKa = 11.84SSSGRR198 pKa = 11.84QPAYY202 pKa = 10.44AVV204 pKa = 3.25
MM1 pKa = 7.29IAGFFFICWRR11 pKa = 11.84IGQLVTLIIPIGILSWFVDD30 pKa = 3.63GFVKK34 pKa = 10.62NNQLTPTYY42 pKa = 9.91ILVLFIVSVLGIFWALDD59 pKa = 3.12TLIRR63 pKa = 11.84HH64 pKa = 5.9SNAKK68 pKa = 9.99RR69 pKa = 11.84SAHH72 pKa = 5.27FVAFVDD78 pKa = 3.95LCFVGSFIAGVYY90 pKa = 6.8QLRR93 pKa = 11.84RR94 pKa = 11.84IANADD99 pKa = 3.37CGNFRR104 pKa = 11.84FDD106 pKa = 4.89PLTLSLGPFGFAGQSANNPLARR128 pKa = 11.84DD129 pKa = 3.84PTKK132 pKa = 10.66VCAMLKK138 pKa = 9.33TSFAFGIMNIGSFFITSILAVLMHH162 pKa = 5.97HH163 pKa = 6.36HH164 pKa = 6.59EE165 pKa = 4.29EE166 pKa = 4.32RR167 pKa = 11.84EE168 pKa = 4.17HH169 pKa = 6.45EE170 pKa = 4.41KK171 pKa = 10.69GSSRR175 pKa = 11.84RR176 pKa = 11.84RR177 pKa = 11.84GSHH180 pKa = 5.08SSHH183 pKa = 6.83RR184 pKa = 11.84GHH186 pKa = 6.32SGSRR190 pKa = 11.84HH191 pKa = 4.98RR192 pKa = 11.84RR193 pKa = 11.84SSSGRR198 pKa = 11.84QPAYY202 pKa = 10.44AVV204 pKa = 3.25
Molecular weight: 22.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4155681 |
39 |
5891 |
451.0 |
50.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.9 ± 0.021 | 1.265 ± 0.01 |
5.545 ± 0.017 | 6.129 ± 0.024 |
3.639 ± 0.017 | 6.754 ± 0.021 |
2.5 ± 0.011 | 5.105 ± 0.018 |
5.002 ± 0.024 | 8.864 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.087 ± 0.008 | 3.954 ± 0.014 |
6.296 ± 0.031 | 4.073 ± 0.019 |
6.446 ± 0.024 | 8.822 ± 0.033 |
5.857 ± 0.014 | 5.802 ± 0.017 |
1.328 ± 0.009 | 2.63 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
