
Hubei polero-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
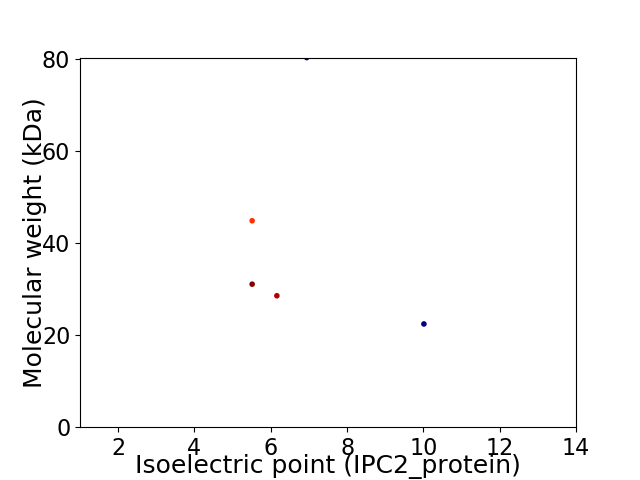
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEP7|A0A1L3KEP7_9VIRU RNA-directed RNA polymerase OS=Hubei polero-like virus 2 OX=1923170 PE=4 SV=1
MM1 pKa = 7.36YY2 pKa = 9.26STVQTSSPDD11 pKa = 3.28LVGYY15 pKa = 8.63TEE17 pKa = 4.46TGHH20 pKa = 7.19LTLSWEE26 pKa = 4.21KK27 pKa = 10.12FQEE30 pKa = 4.09HH31 pKa = 6.18FKK33 pKa = 11.05VAVHH37 pKa = 6.0SLEE40 pKa = 4.04LDD42 pKa = 3.24AGIGIPYY49 pKa = 9.75IKK51 pKa = 10.06YY52 pKa = 9.79RR53 pKa = 11.84RR54 pKa = 11.84PTHH57 pKa = 6.25RR58 pKa = 11.84GWVEE62 pKa = 3.94DD63 pKa = 4.19PEE65 pKa = 4.58LLPVLAQMTFDD76 pKa = 4.44RR77 pKa = 11.84LQKK80 pKa = 9.49MSKK83 pKa = 9.91VFNNNLTPEE92 pKa = 4.12EE93 pKa = 4.24CVQLGLCDD101 pKa = 5.02PIRR104 pKa = 11.84VFIKK108 pKa = 10.73NEE110 pKa = 3.46PHH112 pKa = 6.19KK113 pKa = 10.44QSKK116 pKa = 10.16LDD118 pKa = 3.38EE119 pKa = 4.27GRR121 pKa = 11.84YY122 pKa = 8.96RR123 pKa = 11.84LIMSVSLVDD132 pKa = 3.45QLVARR137 pKa = 11.84ILFQRR142 pKa = 11.84QNKK145 pKa = 8.24MEE147 pKa = 4.34IEE149 pKa = 4.52LWRR152 pKa = 11.84SIPSKK157 pKa = 10.52PGFGLSTDD165 pKa = 3.45EE166 pKa = 4.08QTLEE170 pKa = 4.39FIKK173 pKa = 10.88LLSKK177 pKa = 10.19QANIEE182 pKa = 3.94AAEE185 pKa = 4.45LLTNWADD192 pKa = 3.58YY193 pKa = 10.31VVPTDD198 pKa = 3.9CSGFDD203 pKa = 3.27WSVSQWMLEE212 pKa = 3.9DD213 pKa = 3.35EE214 pKa = 4.43MEE216 pKa = 4.14VRR218 pKa = 11.84NRR220 pKa = 11.84LQSHH224 pKa = 5.95YY225 pKa = 10.62SGHH228 pKa = 5.29TEE230 pKa = 3.63KK231 pKa = 11.02QNFNLQQVWLHH242 pKa = 5.88CLGNSVLALSDD253 pKa = 3.4GRR255 pKa = 11.84LLAQEE260 pKa = 3.94VSGVQKK266 pKa = 10.55SGSYY270 pKa = 8.27NTSSTNSRR278 pKa = 11.84IRR280 pKa = 11.84VMAALHH286 pKa = 6.54CGASWAIAMGDD297 pKa = 3.76DD298 pKa = 4.56AIEE301 pKa = 4.67SNDD304 pKa = 3.71SNLEE308 pKa = 3.88LYY310 pKa = 10.02AQLGFKK316 pKa = 10.75VEE318 pKa = 4.12VSTKK322 pKa = 10.89LEE324 pKa = 4.12FCSHH328 pKa = 6.21IFEE331 pKa = 4.94KK332 pKa = 11.08LNLAIPINKK341 pKa = 9.56GKK343 pKa = 8.85MLYY346 pKa = 10.43KK347 pKa = 10.66LIFGYY352 pKa = 10.7DD353 pKa = 3.51PGCGNVEE360 pKa = 4.12VVTNYY365 pKa = 10.64LNALWSVLHH374 pKa = 6.06EE375 pKa = 4.37LRR377 pKa = 11.84YY378 pKa = 10.78DD379 pKa = 3.75PDD381 pKa = 3.58LVSRR385 pKa = 11.84LHH387 pKa = 6.07KK388 pKa = 10.26WLVPVV393 pKa = 4.26
MM1 pKa = 7.36YY2 pKa = 9.26STVQTSSPDD11 pKa = 3.28LVGYY15 pKa = 8.63TEE17 pKa = 4.46TGHH20 pKa = 7.19LTLSWEE26 pKa = 4.21KK27 pKa = 10.12FQEE30 pKa = 4.09HH31 pKa = 6.18FKK33 pKa = 11.05VAVHH37 pKa = 6.0SLEE40 pKa = 4.04LDD42 pKa = 3.24AGIGIPYY49 pKa = 9.75IKK51 pKa = 10.06YY52 pKa = 9.79RR53 pKa = 11.84RR54 pKa = 11.84PTHH57 pKa = 6.25RR58 pKa = 11.84GWVEE62 pKa = 3.94DD63 pKa = 4.19PEE65 pKa = 4.58LLPVLAQMTFDD76 pKa = 4.44RR77 pKa = 11.84LQKK80 pKa = 9.49MSKK83 pKa = 9.91VFNNNLTPEE92 pKa = 4.12EE93 pKa = 4.24CVQLGLCDD101 pKa = 5.02PIRR104 pKa = 11.84VFIKK108 pKa = 10.73NEE110 pKa = 3.46PHH112 pKa = 6.19KK113 pKa = 10.44QSKK116 pKa = 10.16LDD118 pKa = 3.38EE119 pKa = 4.27GRR121 pKa = 11.84YY122 pKa = 8.96RR123 pKa = 11.84LIMSVSLVDD132 pKa = 3.45QLVARR137 pKa = 11.84ILFQRR142 pKa = 11.84QNKK145 pKa = 8.24MEE147 pKa = 4.34IEE149 pKa = 4.52LWRR152 pKa = 11.84SIPSKK157 pKa = 10.52PGFGLSTDD165 pKa = 3.45EE166 pKa = 4.08QTLEE170 pKa = 4.39FIKK173 pKa = 10.88LLSKK177 pKa = 10.19QANIEE182 pKa = 3.94AAEE185 pKa = 4.45LLTNWADD192 pKa = 3.58YY193 pKa = 10.31VVPTDD198 pKa = 3.9CSGFDD203 pKa = 3.27WSVSQWMLEE212 pKa = 3.9DD213 pKa = 3.35EE214 pKa = 4.43MEE216 pKa = 4.14VRR218 pKa = 11.84NRR220 pKa = 11.84LQSHH224 pKa = 5.95YY225 pKa = 10.62SGHH228 pKa = 5.29TEE230 pKa = 3.63KK231 pKa = 11.02QNFNLQQVWLHH242 pKa = 5.88CLGNSVLALSDD253 pKa = 3.4GRR255 pKa = 11.84LLAQEE260 pKa = 3.94VSGVQKK266 pKa = 10.55SGSYY270 pKa = 8.27NTSSTNSRR278 pKa = 11.84IRR280 pKa = 11.84VMAALHH286 pKa = 6.54CGASWAIAMGDD297 pKa = 3.76DD298 pKa = 4.56AIEE301 pKa = 4.67SNDD304 pKa = 3.71SNLEE308 pKa = 3.88LYY310 pKa = 10.02AQLGFKK316 pKa = 10.75VEE318 pKa = 4.12VSTKK322 pKa = 10.89LEE324 pKa = 4.12FCSHH328 pKa = 6.21IFEE331 pKa = 4.94KK332 pKa = 11.08LNLAIPINKK341 pKa = 9.56GKK343 pKa = 8.85MLYY346 pKa = 10.43KK347 pKa = 10.66LIFGYY352 pKa = 10.7DD353 pKa = 3.51PGCGNVEE360 pKa = 4.12VVTNYY365 pKa = 10.64LNALWSVLHH374 pKa = 6.06EE375 pKa = 4.37LRR377 pKa = 11.84YY378 pKa = 10.78DD379 pKa = 3.75PDD381 pKa = 3.58LVSRR385 pKa = 11.84LHH387 pKa = 6.07KK388 pKa = 10.26WLVPVV393 pKa = 4.26
Molecular weight: 44.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEI1|A0A1L3KEI1_9VIRU Uncharacterized protein OS=Hubei polero-like virus 2 OX=1923170 PE=4 SV=1
MM1 pKa = 6.74NTGGARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.61GRR11 pKa = 11.84NGKK14 pKa = 7.81RR15 pKa = 11.84TRR17 pKa = 11.84TGRR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84NPVRR27 pKa = 11.84PIVMVAPTRR36 pKa = 11.84PTWRR40 pKa = 11.84NRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PTRR47 pKa = 11.84SNRR50 pKa = 11.84GRR52 pKa = 11.84GQLRR56 pKa = 11.84NAGGTSNSEE65 pKa = 3.71TFVFNKK71 pKa = 10.35DD72 pKa = 3.42SLKK75 pKa = 10.93GNSKK79 pKa = 8.85GTITFGPSLSEE90 pKa = 4.12SVALSGGVLKK100 pKa = 10.54AYY102 pKa = 10.29HH103 pKa = 6.77EE104 pKa = 4.24YY105 pKa = 10.75KK106 pKa = 8.49ITKK109 pKa = 8.89VNVRR113 pKa = 11.84FFSEE117 pKa = 4.29SASTAEE123 pKa = 3.84GSIAYY128 pKa = 9.14EE129 pKa = 3.89VDD131 pKa = 3.31PHH133 pKa = 7.13CKK135 pKa = 9.06LTEE138 pKa = 4.42LGSTLRR144 pKa = 11.84KK145 pKa = 8.51FTITKK150 pKa = 10.07AGQATFGASKK160 pKa = 10.57INGLEE165 pKa = 3.83WHH167 pKa = 7.31DD168 pKa = 4.0SSEE171 pKa = 4.33DD172 pKa = 3.39QFKK175 pKa = 10.28IHH177 pKa = 6.23YY178 pKa = 8.62KK179 pKa = 10.77GNGSTTTAGYY189 pKa = 8.81FQIRR193 pKa = 11.84YY194 pKa = 7.38WVSLHH199 pKa = 6.02NPKK202 pKa = 10.6
MM1 pKa = 6.74NTGGARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.61GRR11 pKa = 11.84NGKK14 pKa = 7.81RR15 pKa = 11.84TRR17 pKa = 11.84TGRR20 pKa = 11.84ARR22 pKa = 11.84RR23 pKa = 11.84NPVRR27 pKa = 11.84PIVMVAPTRR36 pKa = 11.84PTWRR40 pKa = 11.84NRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84PTRR47 pKa = 11.84SNRR50 pKa = 11.84GRR52 pKa = 11.84GQLRR56 pKa = 11.84NAGGTSNSEE65 pKa = 3.71TFVFNKK71 pKa = 10.35DD72 pKa = 3.42SLKK75 pKa = 10.93GNSKK79 pKa = 8.85GTITFGPSLSEE90 pKa = 4.12SVALSGGVLKK100 pKa = 10.54AYY102 pKa = 10.29HH103 pKa = 6.77EE104 pKa = 4.24YY105 pKa = 10.75KK106 pKa = 8.49ITKK109 pKa = 8.89VNVRR113 pKa = 11.84FFSEE117 pKa = 4.29SASTAEE123 pKa = 3.84GSIAYY128 pKa = 9.14EE129 pKa = 3.89VDD131 pKa = 3.31PHH133 pKa = 7.13CKK135 pKa = 9.06LTEE138 pKa = 4.42LGSTLRR144 pKa = 11.84KK145 pKa = 8.51FTITKK150 pKa = 10.07AGQATFGASKK160 pKa = 10.57INGLEE165 pKa = 3.83WHH167 pKa = 7.31DD168 pKa = 4.0SSEE171 pKa = 4.33DD172 pKa = 3.39QFKK175 pKa = 10.28IHH177 pKa = 6.23YY178 pKa = 8.62KK179 pKa = 10.77GNGSTTTAGYY189 pKa = 8.81FQIRR193 pKa = 11.84YY194 pKa = 7.38WVSLHH199 pKa = 6.02NPKK202 pKa = 10.6
Molecular weight: 22.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1847 |
202 |
726 |
369.4 |
41.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.793 ± 0.414 | 1.678 ± 0.454 |
4.602 ± 0.796 | 6.172 ± 0.383 |
3.844 ± 0.343 | 6.497 ± 0.633 |
2.382 ± 0.154 | 6.064 ± 0.821 |
5.793 ± 0.662 | 9.258 ± 0.974 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.949 ± 0.23 | 5.143 ± 0.235 |
5.468 ± 0.592 | 3.952 ± 0.471 |
5.522 ± 1.126 | 8.988 ± 0.427 |
6.605 ± 1.318 | 5.468 ± 0.592 |
1.462 ± 0.232 | 3.357 ± 0.458 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
