
Aureobasidium subglaciale (strain EXF-2481) (Aureobasidium pullulans var. subglaciale)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Dothideales; Saccotheciaceae; Aureobasidium; Aureobasidium subglaciale
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
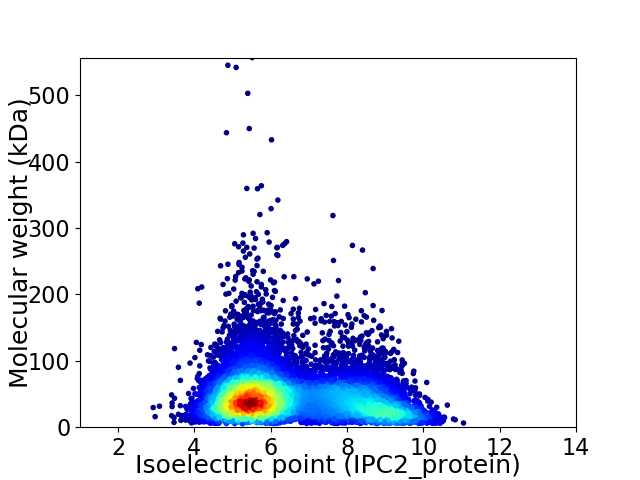
Virtual 2D-PAGE plot for 10792 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
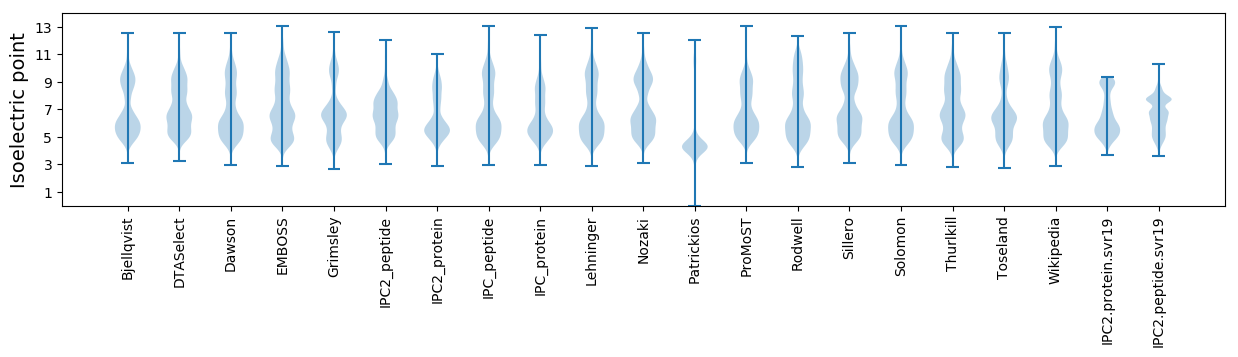
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A074Z0Q4|A0A074Z0Q4_AURSE Atypical serine/threonine protein kinase BUD32 OS=Aureobasidium subglaciale (strain EXF-2481) OX=1043005 GN=AUEXF2481DRAFT_7398 PE=4 SV=1
MM1 pKa = 7.31QKK3 pKa = 10.16RR4 pKa = 11.84SSCTFTSYY12 pKa = 11.75DD13 pKa = 3.81DD14 pKa = 3.94VADD17 pKa = 4.27QIADD21 pKa = 3.77CSSVTLSGITVPAGEE36 pKa = 4.49TLDD39 pKa = 5.08LSDD42 pKa = 4.59AADD45 pKa = 3.86GATITFSGTTTFEE58 pKa = 3.91YY59 pKa = 10.85SEE61 pKa = 4.1WSGPLIRR68 pKa = 11.84AGGKK72 pKa = 9.74GVTITGASGHH82 pKa = 6.25VIDD85 pKa = 5.98GNGAKK90 pKa = 8.86WWDD93 pKa = 3.64GEE95 pKa = 4.32GTNGGKK101 pKa = 8.23TKK103 pKa = 10.58PKK105 pKa = 10.07FFYY108 pKa = 10.56AHH110 pKa = 6.88SLTGNSVIKK119 pKa = 10.46DD120 pKa = 3.72LNIKK124 pKa = 7.56NTPVQAISVQATDD137 pKa = 3.81LTISGLTIDD146 pKa = 5.08NSDD149 pKa = 4.68GDD151 pKa = 5.38AEE153 pKa = 4.54DD154 pKa = 5.38CDD156 pKa = 5.57GEE158 pKa = 4.5ACGHH162 pKa = 5.65NTDD165 pKa = 4.32AFDD168 pKa = 3.47VSEE171 pKa = 4.53SEE173 pKa = 4.13NVVITGCTIKK183 pKa = 11.06NQDD186 pKa = 3.54DD187 pKa = 3.78CLAINSGTGITFSDD201 pKa = 4.0NACSGGHH208 pKa = 6.66GISIGSVGGRR218 pKa = 11.84DD219 pKa = 3.87DD220 pKa = 5.11NDD222 pKa = 3.2VANIIISGNTIKK234 pKa = 10.94DD235 pKa = 3.41SANGLRR241 pKa = 11.84IKK243 pKa = 9.45TVYY246 pKa = 10.47DD247 pKa = 3.17ATGSVKK253 pKa = 10.56NVTYY257 pKa = 10.28TDD259 pKa = 3.49NTISGISDD267 pKa = 3.22YY268 pKa = 11.32GIVVEE273 pKa = 4.61QDD275 pKa = 3.56YY276 pKa = 11.45EE277 pKa = 4.36NGSPTGTPTSGVPITDD293 pKa = 2.77ITFNGVTGTVDD304 pKa = 3.08SDD306 pKa = 3.82AEE308 pKa = 4.16NIYY311 pKa = 10.04ILCASCSGWTWSGVDD326 pKa = 3.11ISGGKK331 pKa = 8.2TSDD334 pKa = 3.02ACEE337 pKa = 4.15GVPSGVSCC345 pKa = 5.13
MM1 pKa = 7.31QKK3 pKa = 10.16RR4 pKa = 11.84SSCTFTSYY12 pKa = 11.75DD13 pKa = 3.81DD14 pKa = 3.94VADD17 pKa = 4.27QIADD21 pKa = 3.77CSSVTLSGITVPAGEE36 pKa = 4.49TLDD39 pKa = 5.08LSDD42 pKa = 4.59AADD45 pKa = 3.86GATITFSGTTTFEE58 pKa = 3.91YY59 pKa = 10.85SEE61 pKa = 4.1WSGPLIRR68 pKa = 11.84AGGKK72 pKa = 9.74GVTITGASGHH82 pKa = 6.25VIDD85 pKa = 5.98GNGAKK90 pKa = 8.86WWDD93 pKa = 3.64GEE95 pKa = 4.32GTNGGKK101 pKa = 8.23TKK103 pKa = 10.58PKK105 pKa = 10.07FFYY108 pKa = 10.56AHH110 pKa = 6.88SLTGNSVIKK119 pKa = 10.46DD120 pKa = 3.72LNIKK124 pKa = 7.56NTPVQAISVQATDD137 pKa = 3.81LTISGLTIDD146 pKa = 5.08NSDD149 pKa = 4.68GDD151 pKa = 5.38AEE153 pKa = 4.54DD154 pKa = 5.38CDD156 pKa = 5.57GEE158 pKa = 4.5ACGHH162 pKa = 5.65NTDD165 pKa = 4.32AFDD168 pKa = 3.47VSEE171 pKa = 4.53SEE173 pKa = 4.13NVVITGCTIKK183 pKa = 11.06NQDD186 pKa = 3.54DD187 pKa = 3.78CLAINSGTGITFSDD201 pKa = 4.0NACSGGHH208 pKa = 6.66GISIGSVGGRR218 pKa = 11.84DD219 pKa = 3.87DD220 pKa = 5.11NDD222 pKa = 3.2VANIIISGNTIKK234 pKa = 10.94DD235 pKa = 3.41SANGLRR241 pKa = 11.84IKK243 pKa = 9.45TVYY246 pKa = 10.47DD247 pKa = 3.17ATGSVKK253 pKa = 10.56NVTYY257 pKa = 10.28TDD259 pKa = 3.49NTISGISDD267 pKa = 3.22YY268 pKa = 11.32GIVVEE273 pKa = 4.61QDD275 pKa = 3.56YY276 pKa = 11.45EE277 pKa = 4.36NGSPTGTPTSGVPITDD293 pKa = 2.77ITFNGVTGTVDD304 pKa = 3.08SDD306 pKa = 3.82AEE308 pKa = 4.16NIYY311 pKa = 10.04ILCASCSGWTWSGVDD326 pKa = 3.11ISGGKK331 pKa = 8.2TSDD334 pKa = 3.02ACEE337 pKa = 4.15GVPSGVSCC345 pKa = 5.13
Molecular weight: 35.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A074YJM6|A0A074YJM6_AURSE Geranylgeranyl transferase type-2 subunit alpha OS=Aureobasidium subglaciale (strain EXF-2481) OX=1043005 GN=AUEXF2481DRAFT_561364 PE=3 SV=1
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 8.59QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.84LAKK15 pKa = 9.55AQKK18 pKa = 8.59QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTIRR36 pKa = 11.84YY37 pKa = 5.73NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4879484 |
49 |
4935 |
452.1 |
50.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.844 ± 0.019 | 1.261 ± 0.009 |
5.776 ± 0.018 | 5.94 ± 0.024 |
3.721 ± 0.015 | 6.558 ± 0.026 |
2.435 ± 0.011 | 4.905 ± 0.016 |
4.939 ± 0.022 | 8.786 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.009 | 3.78 ± 0.012 |
5.825 ± 0.025 | 4.168 ± 0.018 |
5.83 ± 0.019 | 8.457 ± 0.034 |
6.175 ± 0.017 | 6.102 ± 0.015 |
1.423 ± 0.009 | 2.803 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
