
Phytophthora infestans (strain T30-4) (Potato late blight fungus)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora; Phytophthora infestans
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
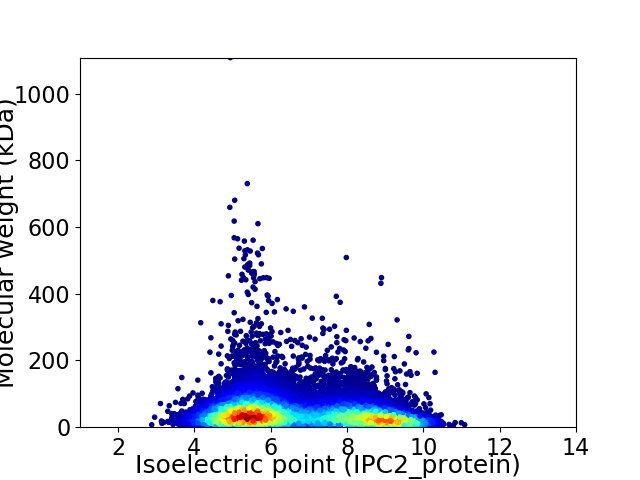
Virtual 2D-PAGE plot for 17610 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
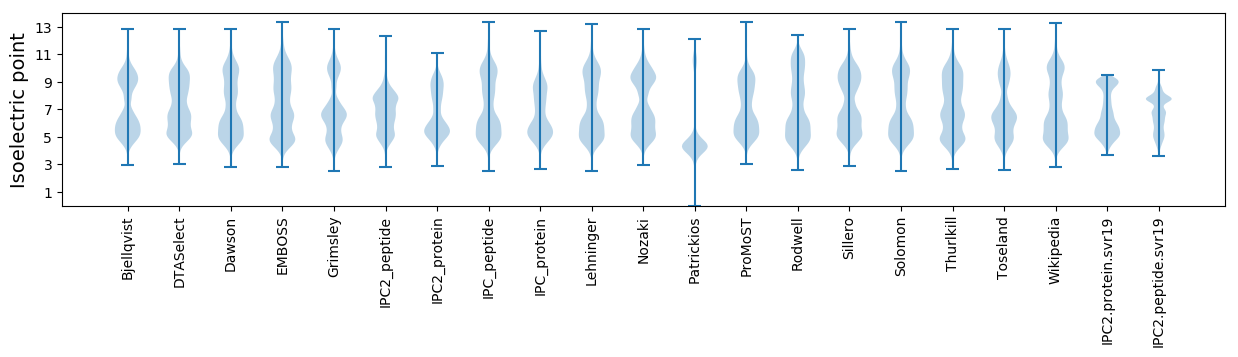
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|D0NM77|D0NM77_PHYIT Secreted RxLR effector peptide protein putative (Fragment) OS=Phytophthora infestans (strain T30-4) OX=403677 GN=PITG_13535 PE=4 SV=1
MM1 pKa = 7.55SPHH4 pKa = 6.67CLSLLVAFGVSFTAAVEE21 pKa = 4.14VSVCRR26 pKa = 11.84DD27 pKa = 2.99ATYY30 pKa = 10.46DD31 pKa = 3.28ISVDD35 pKa = 3.45ATSLCSGSGAAPAGWSCPKK54 pKa = 10.71AGDD57 pKa = 3.7VAVADD62 pKa = 4.52CLSTLASHH70 pKa = 7.36GSGTCVAPEE79 pKa = 3.94DD80 pKa = 4.25AVCQVVNGDD89 pKa = 2.84TWGCVLPSVGCNTPVVADD107 pKa = 4.16SACEE111 pKa = 3.64TWDD114 pKa = 3.42YY115 pKa = 11.83SGDD118 pKa = 3.77DD119 pKa = 3.88TVDD122 pKa = 3.09SSGSFDD128 pKa = 3.57GNEE131 pKa = 4.85DD132 pKa = 3.59YY133 pKa = 11.56DD134 pKa = 4.09EE135 pKa = 4.15TWFMKK140 pKa = 8.33TTEE143 pKa = 4.02LRR145 pKa = 11.84EE146 pKa = 4.62LYY148 pKa = 10.34DD149 pKa = 3.69CSKK152 pKa = 10.83KK153 pKa = 9.0PTPAPTTPCPTPAATEE169 pKa = 4.3TNTTEE174 pKa = 4.42APTATTAPAPTDD186 pKa = 3.52SSDD189 pKa = 3.49TDD191 pKa = 3.87PVTLAPTPTVTPIPMPTEE209 pKa = 4.28VNGSSSDD216 pKa = 3.6VMSDD220 pKa = 2.67GVAQNRR226 pKa = 11.84EE227 pKa = 4.17TEE229 pKa = 4.2VGDD232 pKa = 3.85EE233 pKa = 3.92EE234 pKa = 4.92SAAGTHH240 pKa = 5.14TVVAFAAADD249 pKa = 3.51AVSFGGLSNEE259 pKa = 4.1VVAVIAAVAAFVAVIVAAVAIVYY282 pKa = 9.76ASKK285 pKa = 10.55RR286 pKa = 11.84HH287 pKa = 5.27AKK289 pKa = 10.14EE290 pKa = 3.72DD291 pKa = 3.64VEE293 pKa = 4.47EE294 pKa = 4.98GGEE297 pKa = 3.97KK298 pKa = 10.57DD299 pKa = 3.38EE300 pKa = 5.54SSDD303 pKa = 3.44EE304 pKa = 4.34GEE306 pKa = 4.22EE307 pKa = 4.2KK308 pKa = 10.73EE309 pKa = 4.38GDD311 pKa = 3.63VGEE314 pKa = 4.59DD315 pKa = 3.54AEE317 pKa = 5.55DD318 pKa = 3.63EE319 pKa = 4.65AEE321 pKa = 4.22STTYY325 pKa = 10.16PVAPPTPAVVMGEE338 pKa = 4.06MATTPVAASAKK349 pKa = 9.98AKK351 pKa = 8.74ATSPVTTPAVTSSPEE366 pKa = 3.87ATVEE370 pKa = 4.08TSSDD374 pKa = 3.48HH375 pKa = 6.9VSEE378 pKa = 4.39STDD381 pKa = 2.98GDD383 pKa = 4.02KK384 pKa = 10.77ATEE387 pKa = 4.14TSDD390 pKa = 3.52TPSSQEE396 pKa = 3.95TIAKK400 pKa = 10.17DD401 pKa = 3.46DD402 pKa = 3.56
MM1 pKa = 7.55SPHH4 pKa = 6.67CLSLLVAFGVSFTAAVEE21 pKa = 4.14VSVCRR26 pKa = 11.84DD27 pKa = 2.99ATYY30 pKa = 10.46DD31 pKa = 3.28ISVDD35 pKa = 3.45ATSLCSGSGAAPAGWSCPKK54 pKa = 10.71AGDD57 pKa = 3.7VAVADD62 pKa = 4.52CLSTLASHH70 pKa = 7.36GSGTCVAPEE79 pKa = 3.94DD80 pKa = 4.25AVCQVVNGDD89 pKa = 2.84TWGCVLPSVGCNTPVVADD107 pKa = 4.16SACEE111 pKa = 3.64TWDD114 pKa = 3.42YY115 pKa = 11.83SGDD118 pKa = 3.77DD119 pKa = 3.88TVDD122 pKa = 3.09SSGSFDD128 pKa = 3.57GNEE131 pKa = 4.85DD132 pKa = 3.59YY133 pKa = 11.56DD134 pKa = 4.09EE135 pKa = 4.15TWFMKK140 pKa = 8.33TTEE143 pKa = 4.02LRR145 pKa = 11.84EE146 pKa = 4.62LYY148 pKa = 10.34DD149 pKa = 3.69CSKK152 pKa = 10.83KK153 pKa = 9.0PTPAPTTPCPTPAATEE169 pKa = 4.3TNTTEE174 pKa = 4.42APTATTAPAPTDD186 pKa = 3.52SSDD189 pKa = 3.49TDD191 pKa = 3.87PVTLAPTPTVTPIPMPTEE209 pKa = 4.28VNGSSSDD216 pKa = 3.6VMSDD220 pKa = 2.67GVAQNRR226 pKa = 11.84EE227 pKa = 4.17TEE229 pKa = 4.2VGDD232 pKa = 3.85EE233 pKa = 3.92EE234 pKa = 4.92SAAGTHH240 pKa = 5.14TVVAFAAADD249 pKa = 3.51AVSFGGLSNEE259 pKa = 4.1VVAVIAAVAAFVAVIVAAVAIVYY282 pKa = 9.76ASKK285 pKa = 10.55RR286 pKa = 11.84HH287 pKa = 5.27AKK289 pKa = 10.14EE290 pKa = 3.72DD291 pKa = 3.64VEE293 pKa = 4.47EE294 pKa = 4.98GGEE297 pKa = 3.97KK298 pKa = 10.57DD299 pKa = 3.38EE300 pKa = 5.54SSDD303 pKa = 3.44EE304 pKa = 4.34GEE306 pKa = 4.22EE307 pKa = 4.2KK308 pKa = 10.73EE309 pKa = 4.38GDD311 pKa = 3.63VGEE314 pKa = 4.59DD315 pKa = 3.54AEE317 pKa = 5.55DD318 pKa = 3.63EE319 pKa = 4.65AEE321 pKa = 4.22STTYY325 pKa = 10.16PVAPPTPAVVMGEE338 pKa = 4.06MATTPVAASAKK349 pKa = 9.98AKK351 pKa = 8.74ATSPVTTPAVTSSPEE366 pKa = 3.87ATVEE370 pKa = 4.08TSSDD374 pKa = 3.48HH375 pKa = 6.9VSEE378 pKa = 4.39STDD381 pKa = 2.98GDD383 pKa = 4.02KK384 pKa = 10.77ATEE387 pKa = 4.14TSDD390 pKa = 3.52TPSSQEE396 pKa = 3.95TIAKK400 pKa = 10.17DD401 pKa = 3.46DD402 pKa = 3.56
Molecular weight: 40.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0N7X4|D0N7X4_PHYIT Uncharacterized protein OS=Phytophthora infestans (strain T30-4) OX=403677 GN=PITG_06714 PE=4 SV=1
LL1 pKa = 7.95LLVIWWWQRR10 pKa = 11.84LRR12 pKa = 11.84LLPRR16 pKa = 11.84LLFVVQWWVWQWKK29 pKa = 8.98WILPGLLLFLRR40 pKa = 11.84LPQLRR45 pKa = 11.84RR46 pKa = 11.84LRR48 pKa = 11.84LLQLRR53 pKa = 11.84VPGLLGLL60 pKa = 4.26
LL1 pKa = 7.95LLVIWWWQRR10 pKa = 11.84LRR12 pKa = 11.84LLPRR16 pKa = 11.84LLFVVQWWVWQWKK29 pKa = 8.98WILPGLLLFLRR40 pKa = 11.84LPQLRR45 pKa = 11.84RR46 pKa = 11.84LRR48 pKa = 11.84LLQLRR53 pKa = 11.84VPGLLGLL60 pKa = 4.26
Molecular weight: 7.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7440462 |
29 |
10550 |
422.5 |
46.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.82 ± 0.018 | 1.677 ± 0.011 |
5.63 ± 0.014 | 6.476 ± 0.021 |
3.841 ± 0.011 | 6.12 ± 0.02 |
2.355 ± 0.008 | 4.181 ± 0.013 |
5.212 ± 0.02 | 9.534 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.473 ± 0.009 | 3.576 ± 0.009 |
4.574 ± 0.017 | 4.182 ± 0.014 |
6.269 ± 0.02 | 8.124 ± 0.02 |
5.933 ± 0.019 | 7.113 ± 0.016 |
1.235 ± 0.007 | 2.674 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
