
Thiohalomonas denitrificans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Thiohalomonas
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
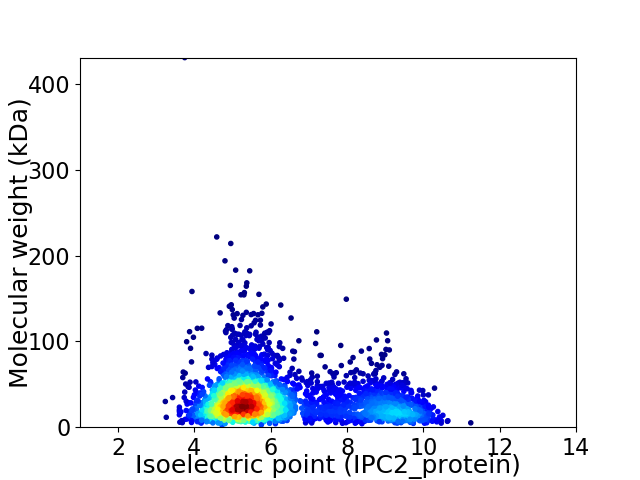
Virtual 2D-PAGE plot for 3255 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G5PQV2|A0A1G5PQV2_9GAMM Osmotically-inducible protein OsmY contains BON domain OS=Thiohalomonas denitrificans OX=415747 GN=SAMN03097708_00599 PE=4 SV=1
MM1 pKa = 6.41MTSLSAQRR9 pKa = 11.84GYY11 pKa = 9.91ATLVVSLILLVAITLITLYY30 pKa = 10.68GARR33 pKa = 11.84TSLMTQRR40 pKa = 11.84ILGNDD45 pKa = 2.96KK46 pKa = 10.41RR47 pKa = 11.84ATEE50 pKa = 4.21SFVEE54 pKa = 4.25AEE56 pKa = 4.1RR57 pKa = 11.84GLDD60 pKa = 3.39EE61 pKa = 4.17GAMFISRR68 pKa = 11.84NIRR71 pKa = 11.84DD72 pKa = 4.14LSNAWVTEE80 pKa = 4.14TRR82 pKa = 11.84WVSCGTSIEE91 pKa = 4.38LPCGDD96 pKa = 3.64GSQNIFDD103 pKa = 4.72ANFVYY108 pKa = 10.87YY109 pKa = 10.02QDD111 pKa = 4.92PDD113 pKa = 4.4DD114 pKa = 3.88NSKK117 pKa = 11.39NYY119 pKa = 9.58VAEE122 pKa = 4.44LSDD125 pKa = 3.88DD126 pKa = 3.74EE127 pKa = 4.29PRR129 pKa = 11.84RR130 pKa = 11.84VYY132 pKa = 10.64LVARR136 pKa = 11.84CQDD139 pKa = 3.53EE140 pKa = 4.67DD141 pKa = 4.61PPDD144 pKa = 4.03GTCDD148 pKa = 3.28HH149 pKa = 6.97VGPGAYY155 pKa = 9.33TIDD158 pKa = 4.66PDD160 pKa = 4.33DD161 pKa = 5.69HH162 pKa = 8.51DD163 pKa = 5.99DD164 pKa = 3.52EE165 pKa = 5.9DD166 pKa = 3.89MRR168 pKa = 11.84GSLIHH173 pKa = 6.77TIATGTSDD181 pKa = 5.1DD182 pKa = 4.04GTGSATIRR190 pKa = 11.84QSHH193 pKa = 6.7YY194 pKa = 10.02IYY196 pKa = 10.61QLVASSPASPLTIDD210 pKa = 3.39AASNVSGSFEE220 pKa = 4.09IVTNPDD226 pKa = 2.81GGGTGVPVSMWSDD239 pKa = 3.42QPIDD243 pKa = 3.51VGTAITTCEE252 pKa = 3.82VGEE255 pKa = 4.23YY256 pKa = 9.98EE257 pKa = 5.46LNDD260 pKa = 3.83GSCPGTTPEE269 pKa = 4.9SLTADD274 pKa = 3.53GDD276 pKa = 3.9AGIDD280 pKa = 3.24IVGEE284 pKa = 4.11ANPFPADD291 pKa = 3.03MFDD294 pKa = 3.79YY295 pKa = 11.04VFGIPTEE302 pKa = 4.38EE303 pKa = 3.93YY304 pKa = 10.92NQVKK308 pKa = 10.18DD309 pKa = 3.27QATAAGHH316 pKa = 7.26LLTDD320 pKa = 4.65CSTLDD325 pKa = 3.4TASNGLYY332 pKa = 9.53WIEE335 pKa = 4.08GACDD339 pKa = 3.46LKK341 pKa = 11.09IEE343 pKa = 4.65IGSSAGPVLLVIADD357 pKa = 3.82GQLDD361 pKa = 3.68INAGAEE367 pKa = 4.38VYY369 pKa = 10.74GLVYY373 pKa = 10.8AFDD376 pKa = 4.19VPPEE380 pKa = 4.77DD381 pKa = 4.1DD382 pKa = 4.82AGDD385 pKa = 3.34IKK387 pKa = 11.4LNGGALIRR395 pKa = 11.84GGFVADD401 pKa = 5.48RR402 pKa = 11.84KK403 pKa = 8.61PTTNGAMTIYY413 pKa = 10.7YY414 pKa = 10.35DD415 pKa = 4.26ADD417 pKa = 3.87LLEE420 pKa = 4.51KK421 pKa = 10.79LVDD424 pKa = 3.93NEE426 pKa = 4.15SLAASGRR433 pKa = 11.84VPGSWVDD440 pKa = 3.2YY441 pKa = 11.26HH442 pKa = 6.27EE443 pKa = 4.39
MM1 pKa = 6.41MTSLSAQRR9 pKa = 11.84GYY11 pKa = 9.91ATLVVSLILLVAITLITLYY30 pKa = 10.68GARR33 pKa = 11.84TSLMTQRR40 pKa = 11.84ILGNDD45 pKa = 2.96KK46 pKa = 10.41RR47 pKa = 11.84ATEE50 pKa = 4.21SFVEE54 pKa = 4.25AEE56 pKa = 4.1RR57 pKa = 11.84GLDD60 pKa = 3.39EE61 pKa = 4.17GAMFISRR68 pKa = 11.84NIRR71 pKa = 11.84DD72 pKa = 4.14LSNAWVTEE80 pKa = 4.14TRR82 pKa = 11.84WVSCGTSIEE91 pKa = 4.38LPCGDD96 pKa = 3.64GSQNIFDD103 pKa = 4.72ANFVYY108 pKa = 10.87YY109 pKa = 10.02QDD111 pKa = 4.92PDD113 pKa = 4.4DD114 pKa = 3.88NSKK117 pKa = 11.39NYY119 pKa = 9.58VAEE122 pKa = 4.44LSDD125 pKa = 3.88DD126 pKa = 3.74EE127 pKa = 4.29PRR129 pKa = 11.84RR130 pKa = 11.84VYY132 pKa = 10.64LVARR136 pKa = 11.84CQDD139 pKa = 3.53EE140 pKa = 4.67DD141 pKa = 4.61PPDD144 pKa = 4.03GTCDD148 pKa = 3.28HH149 pKa = 6.97VGPGAYY155 pKa = 9.33TIDD158 pKa = 4.66PDD160 pKa = 4.33DD161 pKa = 5.69HH162 pKa = 8.51DD163 pKa = 5.99DD164 pKa = 3.52EE165 pKa = 5.9DD166 pKa = 3.89MRR168 pKa = 11.84GSLIHH173 pKa = 6.77TIATGTSDD181 pKa = 5.1DD182 pKa = 4.04GTGSATIRR190 pKa = 11.84QSHH193 pKa = 6.7YY194 pKa = 10.02IYY196 pKa = 10.61QLVASSPASPLTIDD210 pKa = 3.39AASNVSGSFEE220 pKa = 4.09IVTNPDD226 pKa = 2.81GGGTGVPVSMWSDD239 pKa = 3.42QPIDD243 pKa = 3.51VGTAITTCEE252 pKa = 3.82VGEE255 pKa = 4.23YY256 pKa = 9.98EE257 pKa = 5.46LNDD260 pKa = 3.83GSCPGTTPEE269 pKa = 4.9SLTADD274 pKa = 3.53GDD276 pKa = 3.9AGIDD280 pKa = 3.24IVGEE284 pKa = 4.11ANPFPADD291 pKa = 3.03MFDD294 pKa = 3.79YY295 pKa = 11.04VFGIPTEE302 pKa = 4.38EE303 pKa = 3.93YY304 pKa = 10.92NQVKK308 pKa = 10.18DD309 pKa = 3.27QATAAGHH316 pKa = 7.26LLTDD320 pKa = 4.65CSTLDD325 pKa = 3.4TASNGLYY332 pKa = 9.53WIEE335 pKa = 4.08GACDD339 pKa = 3.46LKK341 pKa = 11.09IEE343 pKa = 4.65IGSSAGPVLLVIADD357 pKa = 3.82GQLDD361 pKa = 3.68INAGAEE367 pKa = 4.38VYY369 pKa = 10.74GLVYY373 pKa = 10.8AFDD376 pKa = 4.19VPPEE380 pKa = 4.77DD381 pKa = 4.1DD382 pKa = 4.82AGDD385 pKa = 3.34IKK387 pKa = 11.4LNGGALIRR395 pKa = 11.84GGFVADD401 pKa = 5.48RR402 pKa = 11.84KK403 pKa = 8.61PTTNGAMTIYY413 pKa = 10.7YY414 pKa = 10.35DD415 pKa = 4.26ADD417 pKa = 3.87LLEE420 pKa = 4.51KK421 pKa = 10.79LVDD424 pKa = 3.93NEE426 pKa = 4.15SLAASGRR433 pKa = 11.84VPGSWVDD440 pKa = 3.2YY441 pKa = 11.26HH442 pKa = 6.27EE443 pKa = 4.39
Molecular weight: 47.23 kDa
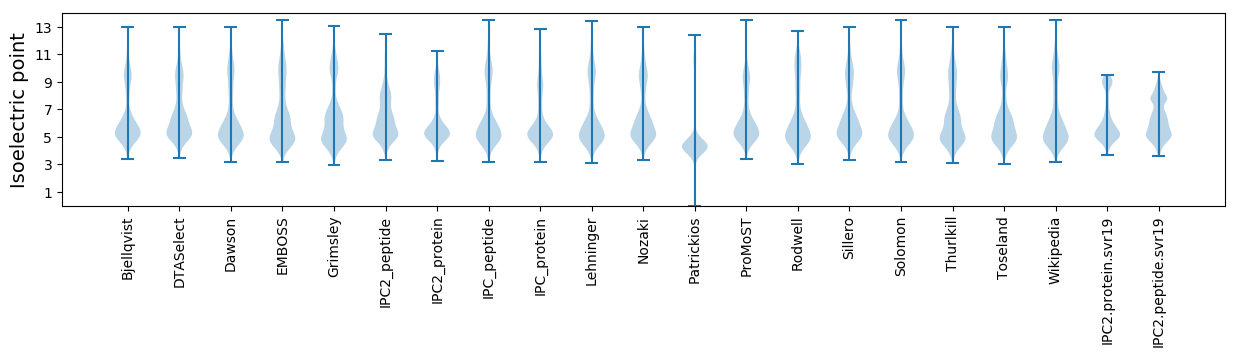
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G5QX78|A0A1G5QX78_9GAMM Uncharacterized protein OS=Thiohalomonas denitrificans OX=415747 GN=SAMN03097708_02889 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.2RR14 pKa = 11.84THH16 pKa = 6.02GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 8.1VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNIRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.2RR14 pKa = 11.84THH16 pKa = 6.02GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTVGGRR28 pKa = 11.84KK29 pKa = 8.1VLKK32 pKa = 10.25ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.31GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1022376 |
24 |
4124 |
314.1 |
34.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.118 ± 0.041 | 0.921 ± 0.016 |
5.61 ± 0.036 | 7.268 ± 0.051 |
3.687 ± 0.029 | 8.211 ± 0.039 |
2.397 ± 0.026 | 4.932 ± 0.032 |
3.242 ± 0.039 | 10.664 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.382 ± 0.021 | 2.739 ± 0.022 |
4.946 ± 0.03 | 3.463 ± 0.026 |
7.583 ± 0.051 | 5.421 ± 0.033 |
5.009 ± 0.043 | 7.409 ± 0.04 |
1.389 ± 0.021 | 2.605 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
