
bacterium F16
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
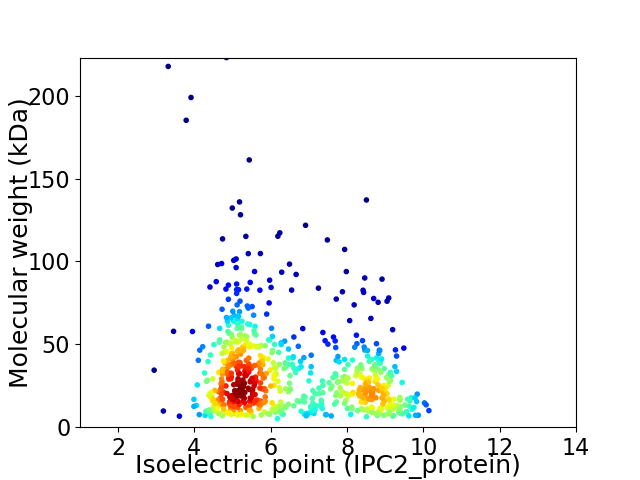
Virtual 2D-PAGE plot for 688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202DNQ0|A0A202DNQ0_9BACT Flagellin OS=bacterium F16 OX=1932694 GN=BVX99_02170 PE=3 SV=1
MM1 pKa = 7.45NGNAGTFSSHH11 pKa = 5.44WLNSLANSDD20 pKa = 3.86GPDD23 pKa = 3.28AVQHH27 pKa = 5.11EE28 pKa = 4.77TAGPGYY34 pKa = 9.59FWNLGPTGIRR44 pKa = 11.84AVLTDD49 pKa = 3.46TNGIIAEE56 pKa = 4.39GVPAGQFIVKK66 pKa = 9.48YY67 pKa = 8.67VFPGSPADD75 pKa = 5.0GIIQVDD81 pKa = 4.32DD82 pKa = 4.68IITGVNAQPFTTSYY96 pKa = 9.66TYY98 pKa = 10.72EE99 pKa = 4.16SNIEE103 pKa = 3.93YY104 pKa = 10.48GFTGPIIAIGNAIEE118 pKa = 4.59ASEE121 pKa = 4.31TNLGGVLTFTVTRR134 pKa = 11.84GGTTSEE140 pKa = 4.0MSVTLDD146 pKa = 3.15NKK148 pKa = 10.01GTFTSTFPYY157 pKa = 10.13SCPKK161 pKa = 10.32SQLLRR166 pKa = 11.84DD167 pKa = 3.83DD168 pKa = 5.63CITEE172 pKa = 4.11LLSLQKK178 pKa = 11.06SNGPWPGGGHH188 pKa = 5.43TSWFALLALMAQGTTHH204 pKa = 7.11LAAVEE209 pKa = 4.24KK210 pKa = 10.78EE211 pKa = 3.87LDD213 pKa = 3.63RR214 pKa = 11.84VSNPVDD220 pKa = 3.54LGTNNWMLAMRR231 pKa = 11.84GIAMAEE237 pKa = 4.03HH238 pKa = 6.17QLLTNSGRR246 pKa = 11.84YY247 pKa = 8.96LDD249 pKa = 4.59AMTAISNQLVANQTKK264 pKa = 9.95YY265 pKa = 10.74GNGAFGHH272 pKa = 6.14SGGLTPGVEE281 pKa = 4.01LGYY284 pKa = 11.26GPMSGITSLVLLSWVMMEE302 pKa = 4.15KK303 pKa = 10.65AGIAVNQVAMKK314 pKa = 8.85KK315 pKa = 7.36TAAVLDD321 pKa = 3.4MRR323 pKa = 11.84LPVNTLGRR331 pKa = 11.84YY332 pKa = 8.52SYY334 pKa = 11.14GWGIQTEE341 pKa = 4.71YY342 pKa = 9.12PTATVDD348 pKa = 3.38RR349 pKa = 11.84PEE351 pKa = 4.25VVASLGDD358 pKa = 3.55LHH360 pKa = 7.96ADD362 pKa = 3.23TATYY366 pKa = 10.62LDD368 pKa = 4.0GVRR371 pKa = 11.84EE372 pKa = 4.02LTIADD377 pKa = 4.64GYY379 pKa = 11.43AGIIHH384 pKa = 6.75SIWPWQPYY392 pKa = 9.08SDD394 pKa = 5.0EE395 pKa = 4.38ISTHH399 pKa = 5.92HH400 pKa = 6.64SNHH403 pKa = 6.51LARR406 pKa = 11.84CRR408 pKa = 11.84RR409 pKa = 11.84VLVNGHH415 pKa = 6.22GSAAMTINGAFLNLIIAGNVGNTSLLSLAMQEE447 pKa = 4.52NKK449 pKa = 10.72ALLNTSRR456 pKa = 11.84CVDD459 pKa = 2.98GGMYY463 pKa = 7.47MQPTRR468 pKa = 11.84DD469 pKa = 3.27HH470 pKa = 7.0SGTDD474 pKa = 3.34LPRR477 pKa = 11.84GSRR480 pKa = 11.84TLATSIWALIFSAPDD495 pKa = 2.88KK496 pKa = 11.07GLYY499 pKa = 10.2LLGRR503 pKa = 11.84GFSDD507 pKa = 4.06AVNEE511 pKa = 4.06APIAYY516 pKa = 8.24YY517 pKa = 10.37QSLQTQEE524 pKa = 3.71ATEE527 pKa = 4.15RR528 pKa = 11.84PITLDD533 pKa = 3.39ANDD536 pKa = 3.92VEE538 pKa = 5.08RR539 pKa = 11.84SSLTYY544 pKa = 10.43SVVSGPSNGIITGSGSSISYY564 pKa = 9.29TPDD567 pKa = 3.25DD568 pKa = 4.46GFAGTDD574 pKa = 3.09SFTFKK579 pKa = 11.17ANDD582 pKa = 3.74GDD584 pKa = 4.25DD585 pKa = 4.06DD586 pKa = 5.73SNIAVVDD593 pKa = 3.16ITVYY597 pKa = 10.58RR598 pKa = 11.84NVFVSVEE605 pKa = 4.02ATDD608 pKa = 4.87DD609 pKa = 3.59VALEE613 pKa = 4.31RR614 pKa = 11.84GTDD617 pKa = 3.21TGAWTVSRR625 pKa = 11.84TGDD628 pKa = 3.66TISPLVIPYY637 pKa = 7.0TTSGTAVGGVDD648 pKa = 4.07YY649 pKa = 10.86LALPDD654 pKa = 4.42SVTIPAGQSSATLTLTPIDD673 pKa = 3.65DD674 pKa = 3.99LVYY677 pKa = 10.77NEE679 pKa = 4.45PDD681 pKa = 3.15EE682 pKa = 4.63TVVLTVTDD690 pKa = 3.88TPEE693 pKa = 4.36LFTIAAGEE701 pKa = 4.04ATVWIDD707 pKa = 5.15DD708 pKa = 4.21NDD710 pKa = 3.99PNAAPTMTFNSPTTDD725 pKa = 3.29VIAIPSTVGLMLDD738 pKa = 3.52VTVTDD743 pKa = 4.94DD744 pKa = 5.51GIPLIPGALTISWTQISGPGTATFEE769 pKa = 4.59SPTAASTAVTFDD781 pKa = 3.67LDD783 pKa = 3.59GTYY786 pKa = 10.08MLTLTADD793 pKa = 4.07DD794 pKa = 4.58SDD796 pKa = 4.0LASTQRR802 pKa = 11.84IVVFVGGVGPGLWYY816 pKa = 10.82GDD818 pKa = 3.23VGGNIDD824 pKa = 4.54LTTPNPKK831 pKa = 9.15TDD833 pKa = 3.48VLVDD837 pKa = 4.01PSSKK841 pKa = 9.85TEE843 pKa = 4.07DD844 pKa = 3.8AIALNTTEE852 pKa = 5.82IYY854 pKa = 10.01TGWIYY859 pKa = 11.13DD860 pKa = 3.44ADD862 pKa = 4.18GNISFTGIIDD872 pKa = 3.69DD873 pKa = 4.19KK874 pKa = 11.43LRR876 pKa = 11.84IFIDD880 pKa = 4.05GALVLSNEE888 pKa = 4.01SRR890 pKa = 11.84NRR892 pKa = 11.84ASTSNLNLTPGWHH905 pKa = 7.26DD906 pKa = 3.07IEE908 pKa = 4.41IRR910 pKa = 11.84ISNGHH915 pKa = 6.17SGSGPINAPGIGYY928 pKa = 10.11DD929 pKa = 4.0PDD931 pKa = 5.8GGTAWQVLTDD941 pKa = 4.28PGDD944 pKa = 3.66GSFLQASFNGNIGPDD959 pKa = 3.4VTATSTSDD967 pKa = 3.25GSNGDD972 pKa = 3.7PMSISATVSDD982 pKa = 5.36DD983 pKa = 3.96NLPTPPAAVATQWLQVPDD1001 pKa = 5.02DD1002 pKa = 4.4GPLTFVDD1009 pKa = 3.56ATSITTEE1016 pKa = 4.24VACMSAATYY1025 pKa = 7.39TLRR1028 pKa = 11.84LIANDD1033 pKa = 4.03GQVTTFDD1040 pKa = 3.51DD1041 pKa = 3.88HH1042 pKa = 6.87GVTFASATRR1051 pKa = 11.84SVSATATDD1059 pKa = 3.48DD1060 pKa = 3.51TAAEE1064 pKa = 4.41TGSEE1068 pKa = 4.09TGTWTITRR1076 pKa = 11.84NGDD1079 pKa = 3.17LSAGLTVYY1087 pKa = 10.82YY1088 pKa = 10.54SLGGTASAADD1098 pKa = 3.77YY1099 pKa = 9.43TVTPLNSVTMAPNAASATVTLTPIDD1124 pKa = 4.27DD1125 pKa = 4.17ALKK1128 pKa = 10.2EE1129 pKa = 4.18GPEE1132 pKa = 4.23TVEE1135 pKa = 4.16LTITPDD1141 pKa = 2.9AAYY1144 pKa = 10.09FSDD1147 pKa = 3.58ATASTLTIVDD1157 pKa = 4.35SDD1159 pKa = 3.78NQAPIVNAGPDD1170 pKa = 3.39QPVQLVAVPLPPGWSSAAWTGDD1192 pKa = 3.12ADD1194 pKa = 4.28SGIDD1198 pKa = 4.04NEE1200 pKa = 4.55LTYY1203 pKa = 10.16TAAHH1207 pKa = 6.45CFGNNHH1213 pKa = 6.91AGTVTVDD1220 pKa = 2.72RR1221 pKa = 11.84VEE1223 pKa = 4.25FTEE1226 pKa = 5.04SFDD1229 pKa = 3.61TTGTGWSITGGKK1241 pKa = 9.82ANWDD1245 pKa = 3.84GNDD1248 pKa = 4.03DD1249 pKa = 5.22AILTDD1254 pKa = 3.77GSEE1257 pKa = 4.24DD1258 pKa = 3.36LGEE1261 pKa = 3.96EE1262 pKa = 4.77FIYY1265 pKa = 10.63GGNPRR1270 pKa = 11.84TVTFTGLTPGDD1281 pKa = 4.24SYY1283 pKa = 11.48TATFFSVGWDD1293 pKa = 3.44TTASRR1298 pKa = 11.84EE1299 pKa = 4.02QTFSITGGSSLLVDD1313 pKa = 3.37QNSYY1317 pKa = 9.28GKK1319 pKa = 10.19NYY1321 pKa = 9.85GISVSCNYY1329 pKa = 8.19TATATSQEE1337 pKa = 4.31VVIAPVAGSFHH1348 pKa = 7.26LYY1350 pKa = 10.94AMANADD1356 pKa = 3.88TNMLQATAPLSGTVTDD1372 pKa = 5.43DD1373 pKa = 5.61DD1374 pKa = 5.35GDD1376 pKa = 3.93TPSAIWSLVDD1386 pKa = 3.87GPAPVTFDD1394 pKa = 4.21DD1395 pKa = 4.32ASAMITNAYY1404 pKa = 7.51FTEE1407 pKa = 4.64AGTYY1411 pKa = 7.09TLRR1414 pKa = 11.84LTADD1418 pKa = 3.52DD1419 pKa = 4.27TFEE1422 pKa = 4.03PAYY1425 pKa = 10.27DD1426 pKa = 3.61DD1427 pKa = 4.14VIIIVGTGNEE1437 pKa = 4.41IYY1439 pKa = 10.71SVTYY1443 pKa = 10.49DD1444 pKa = 3.31GNTNDD1449 pKa = 4.6GGAIPTGSVDD1459 pKa = 3.43YY1460 pKa = 11.07NFNEE1464 pKa = 4.28VVTVLDD1470 pKa = 3.43NTGNLTKK1477 pKa = 10.16TGYY1480 pKa = 8.67TFTGWNTAANGSGAIYY1496 pKa = 8.95TAGDD1500 pKa = 3.76TFVITADD1507 pKa = 3.52TVLYY1511 pKa = 9.42AQWSINSYY1519 pKa = 10.86AVTYY1523 pKa = 10.37DD1524 pKa = 3.68GNSADD1529 pKa = 4.11NGTVPAVQTKK1539 pKa = 10.21EE1540 pKa = 3.78YY1541 pKa = 10.53GADD1544 pKa = 2.73ITLAFNTGNLSQSGLTFAGWNTASDD1569 pKa = 3.95DD1570 pKa = 4.08SGTHH1574 pKa = 4.59YY1575 pKa = 11.36AEE1577 pKa = 4.43GATYY1581 pKa = 10.88SDD1583 pKa = 3.69NTDD1586 pKa = 2.98LTLYY1590 pKa = 10.61AEE1592 pKa = 4.49WSSVPVYY1599 pKa = 11.01SLTVNNGTGTGLYY1612 pKa = 8.16EE1613 pKa = 4.56ANDD1616 pKa = 3.82VVSVEE1621 pKa = 3.99ASIPANYY1628 pKa = 10.12HH1629 pKa = 4.5FVNWSGDD1636 pKa = 3.25TAAVGDD1642 pKa = 4.24VNSSSTMVTMPAADD1656 pKa = 3.49VTITANVAIDD1666 pKa = 3.88TYY1668 pKa = 11.6AVTFVEE1674 pKa = 4.95GANGTRR1680 pKa = 11.84TGGGALVQTIDD1691 pKa = 3.6YY1692 pKa = 9.92GSAATAPVITADD1704 pKa = 3.41SGYY1707 pKa = 8.85TFTGWDD1713 pKa = 2.96IAFEE1717 pKa = 4.24NVTSNLTVAAQYY1729 pKa = 11.15SVDD1732 pKa = 3.57AVGGSSGGGGGGGGGCQMGVVTGGSLLPFALLLVVAGFIRR1772 pKa = 11.84RR1773 pKa = 11.84RR1774 pKa = 11.84FF1775 pKa = 3.4
MM1 pKa = 7.45NGNAGTFSSHH11 pKa = 5.44WLNSLANSDD20 pKa = 3.86GPDD23 pKa = 3.28AVQHH27 pKa = 5.11EE28 pKa = 4.77TAGPGYY34 pKa = 9.59FWNLGPTGIRR44 pKa = 11.84AVLTDD49 pKa = 3.46TNGIIAEE56 pKa = 4.39GVPAGQFIVKK66 pKa = 9.48YY67 pKa = 8.67VFPGSPADD75 pKa = 5.0GIIQVDD81 pKa = 4.32DD82 pKa = 4.68IITGVNAQPFTTSYY96 pKa = 9.66TYY98 pKa = 10.72EE99 pKa = 4.16SNIEE103 pKa = 3.93YY104 pKa = 10.48GFTGPIIAIGNAIEE118 pKa = 4.59ASEE121 pKa = 4.31TNLGGVLTFTVTRR134 pKa = 11.84GGTTSEE140 pKa = 4.0MSVTLDD146 pKa = 3.15NKK148 pKa = 10.01GTFTSTFPYY157 pKa = 10.13SCPKK161 pKa = 10.32SQLLRR166 pKa = 11.84DD167 pKa = 3.83DD168 pKa = 5.63CITEE172 pKa = 4.11LLSLQKK178 pKa = 11.06SNGPWPGGGHH188 pKa = 5.43TSWFALLALMAQGTTHH204 pKa = 7.11LAAVEE209 pKa = 4.24KK210 pKa = 10.78EE211 pKa = 3.87LDD213 pKa = 3.63RR214 pKa = 11.84VSNPVDD220 pKa = 3.54LGTNNWMLAMRR231 pKa = 11.84GIAMAEE237 pKa = 4.03HH238 pKa = 6.17QLLTNSGRR246 pKa = 11.84YY247 pKa = 8.96LDD249 pKa = 4.59AMTAISNQLVANQTKK264 pKa = 9.95YY265 pKa = 10.74GNGAFGHH272 pKa = 6.14SGGLTPGVEE281 pKa = 4.01LGYY284 pKa = 11.26GPMSGITSLVLLSWVMMEE302 pKa = 4.15KK303 pKa = 10.65AGIAVNQVAMKK314 pKa = 8.85KK315 pKa = 7.36TAAVLDD321 pKa = 3.4MRR323 pKa = 11.84LPVNTLGRR331 pKa = 11.84YY332 pKa = 8.52SYY334 pKa = 11.14GWGIQTEE341 pKa = 4.71YY342 pKa = 9.12PTATVDD348 pKa = 3.38RR349 pKa = 11.84PEE351 pKa = 4.25VVASLGDD358 pKa = 3.55LHH360 pKa = 7.96ADD362 pKa = 3.23TATYY366 pKa = 10.62LDD368 pKa = 4.0GVRR371 pKa = 11.84EE372 pKa = 4.02LTIADD377 pKa = 4.64GYY379 pKa = 11.43AGIIHH384 pKa = 6.75SIWPWQPYY392 pKa = 9.08SDD394 pKa = 5.0EE395 pKa = 4.38ISTHH399 pKa = 5.92HH400 pKa = 6.64SNHH403 pKa = 6.51LARR406 pKa = 11.84CRR408 pKa = 11.84RR409 pKa = 11.84VLVNGHH415 pKa = 6.22GSAAMTINGAFLNLIIAGNVGNTSLLSLAMQEE447 pKa = 4.52NKK449 pKa = 10.72ALLNTSRR456 pKa = 11.84CVDD459 pKa = 2.98GGMYY463 pKa = 7.47MQPTRR468 pKa = 11.84DD469 pKa = 3.27HH470 pKa = 7.0SGTDD474 pKa = 3.34LPRR477 pKa = 11.84GSRR480 pKa = 11.84TLATSIWALIFSAPDD495 pKa = 2.88KK496 pKa = 11.07GLYY499 pKa = 10.2LLGRR503 pKa = 11.84GFSDD507 pKa = 4.06AVNEE511 pKa = 4.06APIAYY516 pKa = 8.24YY517 pKa = 10.37QSLQTQEE524 pKa = 3.71ATEE527 pKa = 4.15RR528 pKa = 11.84PITLDD533 pKa = 3.39ANDD536 pKa = 3.92VEE538 pKa = 5.08RR539 pKa = 11.84SSLTYY544 pKa = 10.43SVVSGPSNGIITGSGSSISYY564 pKa = 9.29TPDD567 pKa = 3.25DD568 pKa = 4.46GFAGTDD574 pKa = 3.09SFTFKK579 pKa = 11.17ANDD582 pKa = 3.74GDD584 pKa = 4.25DD585 pKa = 4.06DD586 pKa = 5.73SNIAVVDD593 pKa = 3.16ITVYY597 pKa = 10.58RR598 pKa = 11.84NVFVSVEE605 pKa = 4.02ATDD608 pKa = 4.87DD609 pKa = 3.59VALEE613 pKa = 4.31RR614 pKa = 11.84GTDD617 pKa = 3.21TGAWTVSRR625 pKa = 11.84TGDD628 pKa = 3.66TISPLVIPYY637 pKa = 7.0TTSGTAVGGVDD648 pKa = 4.07YY649 pKa = 10.86LALPDD654 pKa = 4.42SVTIPAGQSSATLTLTPIDD673 pKa = 3.65DD674 pKa = 3.99LVYY677 pKa = 10.77NEE679 pKa = 4.45PDD681 pKa = 3.15EE682 pKa = 4.63TVVLTVTDD690 pKa = 3.88TPEE693 pKa = 4.36LFTIAAGEE701 pKa = 4.04ATVWIDD707 pKa = 5.15DD708 pKa = 4.21NDD710 pKa = 3.99PNAAPTMTFNSPTTDD725 pKa = 3.29VIAIPSTVGLMLDD738 pKa = 3.52VTVTDD743 pKa = 4.94DD744 pKa = 5.51GIPLIPGALTISWTQISGPGTATFEE769 pKa = 4.59SPTAASTAVTFDD781 pKa = 3.67LDD783 pKa = 3.59GTYY786 pKa = 10.08MLTLTADD793 pKa = 4.07DD794 pKa = 4.58SDD796 pKa = 4.0LASTQRR802 pKa = 11.84IVVFVGGVGPGLWYY816 pKa = 10.82GDD818 pKa = 3.23VGGNIDD824 pKa = 4.54LTTPNPKK831 pKa = 9.15TDD833 pKa = 3.48VLVDD837 pKa = 4.01PSSKK841 pKa = 9.85TEE843 pKa = 4.07DD844 pKa = 3.8AIALNTTEE852 pKa = 5.82IYY854 pKa = 10.01TGWIYY859 pKa = 11.13DD860 pKa = 3.44ADD862 pKa = 4.18GNISFTGIIDD872 pKa = 3.69DD873 pKa = 4.19KK874 pKa = 11.43LRR876 pKa = 11.84IFIDD880 pKa = 4.05GALVLSNEE888 pKa = 4.01SRR890 pKa = 11.84NRR892 pKa = 11.84ASTSNLNLTPGWHH905 pKa = 7.26DD906 pKa = 3.07IEE908 pKa = 4.41IRR910 pKa = 11.84ISNGHH915 pKa = 6.17SGSGPINAPGIGYY928 pKa = 10.11DD929 pKa = 4.0PDD931 pKa = 5.8GGTAWQVLTDD941 pKa = 4.28PGDD944 pKa = 3.66GSFLQASFNGNIGPDD959 pKa = 3.4VTATSTSDD967 pKa = 3.25GSNGDD972 pKa = 3.7PMSISATVSDD982 pKa = 5.36DD983 pKa = 3.96NLPTPPAAVATQWLQVPDD1001 pKa = 5.02DD1002 pKa = 4.4GPLTFVDD1009 pKa = 3.56ATSITTEE1016 pKa = 4.24VACMSAATYY1025 pKa = 7.39TLRR1028 pKa = 11.84LIANDD1033 pKa = 4.03GQVTTFDD1040 pKa = 3.51DD1041 pKa = 3.88HH1042 pKa = 6.87GVTFASATRR1051 pKa = 11.84SVSATATDD1059 pKa = 3.48DD1060 pKa = 3.51TAAEE1064 pKa = 4.41TGSEE1068 pKa = 4.09TGTWTITRR1076 pKa = 11.84NGDD1079 pKa = 3.17LSAGLTVYY1087 pKa = 10.82YY1088 pKa = 10.54SLGGTASAADD1098 pKa = 3.77YY1099 pKa = 9.43TVTPLNSVTMAPNAASATVTLTPIDD1124 pKa = 4.27DD1125 pKa = 4.17ALKK1128 pKa = 10.2EE1129 pKa = 4.18GPEE1132 pKa = 4.23TVEE1135 pKa = 4.16LTITPDD1141 pKa = 2.9AAYY1144 pKa = 10.09FSDD1147 pKa = 3.58ATASTLTIVDD1157 pKa = 4.35SDD1159 pKa = 3.78NQAPIVNAGPDD1170 pKa = 3.39QPVQLVAVPLPPGWSSAAWTGDD1192 pKa = 3.12ADD1194 pKa = 4.28SGIDD1198 pKa = 4.04NEE1200 pKa = 4.55LTYY1203 pKa = 10.16TAAHH1207 pKa = 6.45CFGNNHH1213 pKa = 6.91AGTVTVDD1220 pKa = 2.72RR1221 pKa = 11.84VEE1223 pKa = 4.25FTEE1226 pKa = 5.04SFDD1229 pKa = 3.61TTGTGWSITGGKK1241 pKa = 9.82ANWDD1245 pKa = 3.84GNDD1248 pKa = 4.03DD1249 pKa = 5.22AILTDD1254 pKa = 3.77GSEE1257 pKa = 4.24DD1258 pKa = 3.36LGEE1261 pKa = 3.96EE1262 pKa = 4.77FIYY1265 pKa = 10.63GGNPRR1270 pKa = 11.84TVTFTGLTPGDD1281 pKa = 4.24SYY1283 pKa = 11.48TATFFSVGWDD1293 pKa = 3.44TTASRR1298 pKa = 11.84EE1299 pKa = 4.02QTFSITGGSSLLVDD1313 pKa = 3.37QNSYY1317 pKa = 9.28GKK1319 pKa = 10.19NYY1321 pKa = 9.85GISVSCNYY1329 pKa = 8.19TATATSQEE1337 pKa = 4.31VVIAPVAGSFHH1348 pKa = 7.26LYY1350 pKa = 10.94AMANADD1356 pKa = 3.88TNMLQATAPLSGTVTDD1372 pKa = 5.43DD1373 pKa = 5.61DD1374 pKa = 5.35GDD1376 pKa = 3.93TPSAIWSLVDD1386 pKa = 3.87GPAPVTFDD1394 pKa = 4.21DD1395 pKa = 4.32ASAMITNAYY1404 pKa = 7.51FTEE1407 pKa = 4.64AGTYY1411 pKa = 7.09TLRR1414 pKa = 11.84LTADD1418 pKa = 3.52DD1419 pKa = 4.27TFEE1422 pKa = 4.03PAYY1425 pKa = 10.27DD1426 pKa = 3.61DD1427 pKa = 4.14VIIIVGTGNEE1437 pKa = 4.41IYY1439 pKa = 10.71SVTYY1443 pKa = 10.49DD1444 pKa = 3.31GNTNDD1449 pKa = 4.6GGAIPTGSVDD1459 pKa = 3.43YY1460 pKa = 11.07NFNEE1464 pKa = 4.28VVTVLDD1470 pKa = 3.43NTGNLTKK1477 pKa = 10.16TGYY1480 pKa = 8.67TFTGWNTAANGSGAIYY1496 pKa = 8.95TAGDD1500 pKa = 3.76TFVITADD1507 pKa = 3.52TVLYY1511 pKa = 9.42AQWSINSYY1519 pKa = 10.86AVTYY1523 pKa = 10.37DD1524 pKa = 3.68GNSADD1529 pKa = 4.11NGTVPAVQTKK1539 pKa = 10.21EE1540 pKa = 3.78YY1541 pKa = 10.53GADD1544 pKa = 2.73ITLAFNTGNLSQSGLTFAGWNTASDD1569 pKa = 3.95DD1570 pKa = 4.08SGTHH1574 pKa = 4.59YY1575 pKa = 11.36AEE1577 pKa = 4.43GATYY1581 pKa = 10.88SDD1583 pKa = 3.69NTDD1586 pKa = 2.98LTLYY1590 pKa = 10.61AEE1592 pKa = 4.49WSSVPVYY1599 pKa = 11.01SLTVNNGTGTGLYY1612 pKa = 8.16EE1613 pKa = 4.56ANDD1616 pKa = 3.82VVSVEE1621 pKa = 3.99ASIPANYY1628 pKa = 10.12HH1629 pKa = 4.5FVNWSGDD1636 pKa = 3.25TAAVGDD1642 pKa = 4.24VNSSSTMVTMPAADD1656 pKa = 3.49VTITANVAIDD1666 pKa = 3.88TYY1668 pKa = 11.6AVTFVEE1674 pKa = 4.95GANGTRR1680 pKa = 11.84TGGGALVQTIDD1691 pKa = 3.6YY1692 pKa = 9.92GSAATAPVITADD1704 pKa = 3.41SGYY1707 pKa = 8.85TFTGWDD1713 pKa = 2.96IAFEE1717 pKa = 4.24NVTSNLTVAAQYY1729 pKa = 11.15SVDD1732 pKa = 3.57AVGGSSGGGGGGGGGCQMGVVTGGSLLPFALLLVVAGFIRR1772 pKa = 11.84RR1773 pKa = 11.84RR1774 pKa = 11.84FF1775 pKa = 3.4
Molecular weight: 185.17 kDa
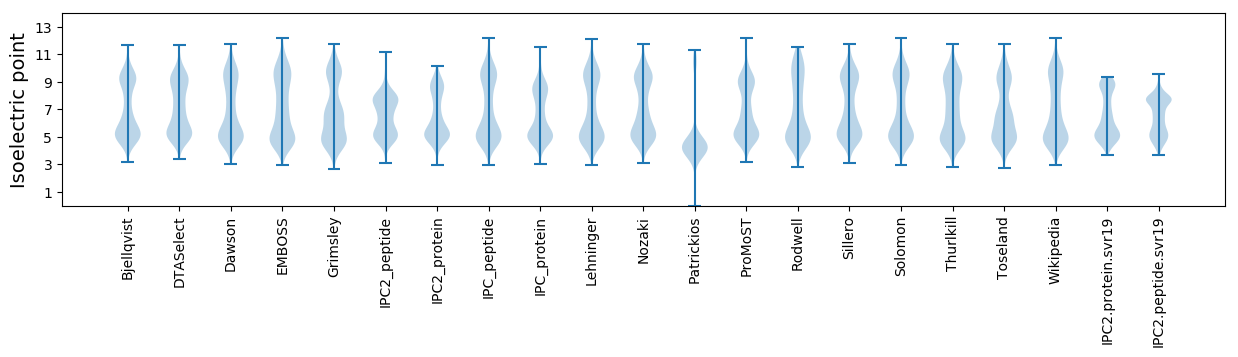
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DPI5|A0A202DPI5_9BACT Uncharacterized protein OS=bacterium F16 OX=1932694 GN=BVX99_01260 PE=4 SV=1
MM1 pKa = 7.19HH2 pKa = 7.64ANMAEE7 pKa = 4.53AIVMADD13 pKa = 3.52VLSGQALGRR22 pKa = 11.84RR23 pKa = 11.84AFEE26 pKa = 3.95TLFVAEE32 pKa = 4.71VGSSPMNHH40 pKa = 6.16LRR42 pKa = 11.84SIRR45 pKa = 11.84LRR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84YY51 pKa = 8.29LLRR54 pKa = 11.84TTDD57 pKa = 3.63LTVKK61 pKa = 10.52EE62 pKa = 4.66IGRR65 pKa = 11.84QCGIPNITYY74 pKa = 10.19LCRR77 pKa = 11.84LFQKK81 pKa = 10.1IHH83 pKa = 5.79GVTPKK88 pKa = 10.23QYY90 pKa = 10.28RR91 pKa = 11.84EE92 pKa = 3.82QRR94 pKa = 3.37
MM1 pKa = 7.19HH2 pKa = 7.64ANMAEE7 pKa = 4.53AIVMADD13 pKa = 3.52VLSGQALGRR22 pKa = 11.84RR23 pKa = 11.84AFEE26 pKa = 3.95TLFVAEE32 pKa = 4.71VGSSPMNHH40 pKa = 6.16LRR42 pKa = 11.84SIRR45 pKa = 11.84LRR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84YY51 pKa = 8.29LLRR54 pKa = 11.84TTDD57 pKa = 3.63LTVKK61 pKa = 10.52EE62 pKa = 4.66IGRR65 pKa = 11.84QCGIPNITYY74 pKa = 10.19LCRR77 pKa = 11.84LFQKK81 pKa = 10.1IHH83 pKa = 5.79GVTPKK88 pKa = 10.23QYY90 pKa = 10.28RR91 pKa = 11.84EE92 pKa = 3.82QRR94 pKa = 3.37
Molecular weight: 10.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
212882 |
46 |
2165 |
309.4 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.882 ± 0.103 | 1.364 ± 0.042 |
6.194 ± 0.071 | 6.263 ± 0.091 |
4.035 ± 0.062 | 7.086 ± 0.098 |
2.238 ± 0.046 | 6.443 ± 0.067 |
5.61 ± 0.119 | 9.721 ± 0.118 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.045 | 4.129 ± 0.054 |
4.473 ± 0.077 | 3.447 ± 0.064 |
5.354 ± 0.081 | 6.479 ± 0.085 |
5.948 ± 0.149 | 6.468 ± 0.08 |
1.234 ± 0.04 | 3.055 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
