
uncultured archaeon A07HR67
Taxonomy: cellular organisms; Archaea; environmental samples
Average proteome isoelectric point is 5.02
Get precalculated fractions of proteins
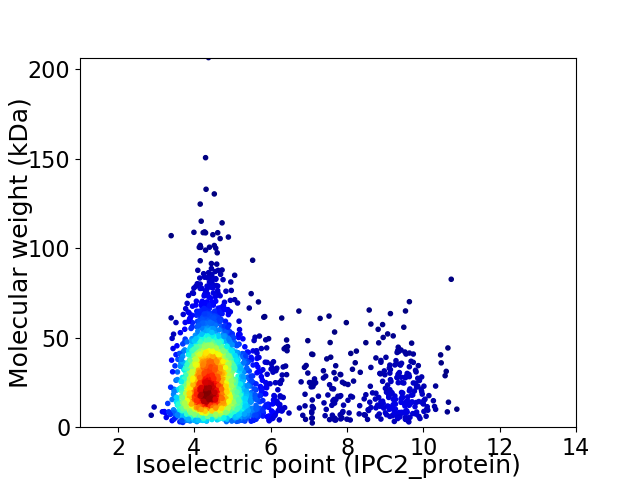
Virtual 2D-PAGE plot for 2887 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V4XS26|V4XS26_9ARCH ATPase involved in chromosome partitioning OS=uncultured archaeon A07HR67 OX=1412871 GN=A07HR67_02377 PE=4 SV=1
MM1 pKa = 7.51AVLVIAIMASAAGNIMEE18 pKa = 4.35QTLGVPYY25 pKa = 10.17LAGVGAVIAVVGVLSYY41 pKa = 11.37VGDD44 pKa = 3.89SLIEE48 pKa = 3.94RR49 pKa = 11.84FKK51 pKa = 10.82TGGTVFLYY59 pKa = 10.55LAYY62 pKa = 9.11LTFGIVVLAATWGDD76 pKa = 3.26ITSVFAAGDD85 pKa = 3.3TSYY88 pKa = 11.83ASDD91 pKa = 3.9ASPLAVLRR99 pKa = 11.84SGILYY104 pKa = 9.9VGYY107 pKa = 10.55NLVVLPAVFFALHH120 pKa = 5.64RR121 pKa = 11.84QVDD124 pKa = 3.6RR125 pKa = 11.84RR126 pKa = 11.84EE127 pKa = 4.24SVVSGVLAGTLMTIPFAITYY147 pKa = 7.47VAFMGFYY154 pKa = 9.84PAEE157 pKa = 4.2NVMGAPVPWLPVLNGVGGPALIGLYY182 pKa = 9.76GVVMGWTLIEE192 pKa = 4.08TSVGLIHH199 pKa = 7.53AIISRR204 pKa = 11.84LDD206 pKa = 3.52TNIEE210 pKa = 4.14EE211 pKa = 4.16IDD213 pKa = 3.48AGPFEE218 pKa = 4.59TQTGLSRR225 pKa = 11.84TQRR228 pKa = 11.84GGLAVGVLVGALGLSRR244 pKa = 11.84VGIIDD249 pKa = 4.84LVAQGYY255 pKa = 5.76TTMAYY260 pKa = 9.92FFIGIFAIPLXXXXXXXPGQLFNFASMYY288 pKa = 10.4VPSNDD293 pKa = 3.16VFVSSAAGIEE303 pKa = 4.26LFNGEE308 pKa = 4.78DD309 pKa = 3.66PVSGEE314 pKa = 3.94VGEE317 pKa = 6.43DD318 pKa = 2.93IALWDD323 pKa = 4.44AGTEE327 pKa = 4.24RR328 pKa = 11.84NGQPGVDD335 pKa = 3.62PVSAPLQAQNGGPMAGPQEE354 pKa = 4.53GVVHH358 pKa = 6.99RR359 pKa = 11.84LDD361 pKa = 5.55DD362 pKa = 4.03INDD365 pKa = 3.68GFDD368 pKa = 3.24YY369 pKa = 11.28ADD371 pKa = 3.45ASEE374 pKa = 4.48IVDD377 pKa = 3.38VTLTHH382 pKa = 7.26DD383 pKa = 3.72GDD385 pKa = 3.97GSFTVRR391 pKa = 11.84LEE393 pKa = 4.1NTSPTDD399 pKa = 3.37VYY401 pKa = 11.5GDD403 pKa = 3.62DD404 pKa = 3.88TATGGQVWITPGAWAVHH421 pKa = 5.93TGSNPIFEE429 pKa = 4.85RR430 pKa = 11.84GEE432 pKa = 3.77PASIGLEE439 pKa = 3.91ALAEE443 pKa = 4.44AGPPTGFEE451 pKa = 4.15GQPGLVDD458 pKa = 4.23EE459 pKa = 5.58LPEE462 pKa = 5.43ADD464 pKa = 3.37NVVDD468 pKa = 3.9VGAYY472 pKa = 7.68TPSNTVTDD480 pKa = 4.59PNDD483 pKa = 3.24PMGAVPGAPPIAPGGAFEE501 pKa = 4.61FTVEE505 pKa = 4.0ADD507 pKa = 3.25PSEE510 pKa = 4.26RR511 pKa = 11.84LSFASMYY518 pKa = 10.48VPSNDD523 pKa = 2.84VFIAPEE529 pKa = 3.92PAIEE533 pKa = 5.19LYY535 pKa = 10.29TDD537 pKa = 3.61DD538 pKa = 4.82GEE540 pKa = 4.33PVEE543 pKa = 4.97GGVTEE548 pKa = 5.86SVGLWDD554 pKa = 5.93AGTEE558 pKa = 4.2LNGRR562 pKa = 11.84PGVDD566 pKa = 3.25PASAPLQAQNGGPTVGPQEE585 pKa = 4.46GVVHH589 pKa = 6.95RR590 pKa = 11.84LDD592 pKa = 6.01DD593 pKa = 3.97IDD595 pKa = 6.33DD596 pKa = 4.33GFDD599 pKa = 3.27YY600 pKa = 11.38ADD602 pKa = 3.47ASEE605 pKa = 4.39IVSVTIEE612 pKa = 3.96PQQ614 pKa = 2.82
MM1 pKa = 7.51AVLVIAIMASAAGNIMEE18 pKa = 4.35QTLGVPYY25 pKa = 10.17LAGVGAVIAVVGVLSYY41 pKa = 11.37VGDD44 pKa = 3.89SLIEE48 pKa = 3.94RR49 pKa = 11.84FKK51 pKa = 10.82TGGTVFLYY59 pKa = 10.55LAYY62 pKa = 9.11LTFGIVVLAATWGDD76 pKa = 3.26ITSVFAAGDD85 pKa = 3.3TSYY88 pKa = 11.83ASDD91 pKa = 3.9ASPLAVLRR99 pKa = 11.84SGILYY104 pKa = 9.9VGYY107 pKa = 10.55NLVVLPAVFFALHH120 pKa = 5.64RR121 pKa = 11.84QVDD124 pKa = 3.6RR125 pKa = 11.84RR126 pKa = 11.84EE127 pKa = 4.24SVVSGVLAGTLMTIPFAITYY147 pKa = 7.47VAFMGFYY154 pKa = 9.84PAEE157 pKa = 4.2NVMGAPVPWLPVLNGVGGPALIGLYY182 pKa = 9.76GVVMGWTLIEE192 pKa = 4.08TSVGLIHH199 pKa = 7.53AIISRR204 pKa = 11.84LDD206 pKa = 3.52TNIEE210 pKa = 4.14EE211 pKa = 4.16IDD213 pKa = 3.48AGPFEE218 pKa = 4.59TQTGLSRR225 pKa = 11.84TQRR228 pKa = 11.84GGLAVGVLVGALGLSRR244 pKa = 11.84VGIIDD249 pKa = 4.84LVAQGYY255 pKa = 5.76TTMAYY260 pKa = 9.92FFIGIFAIPLXXXXXXXPGQLFNFASMYY288 pKa = 10.4VPSNDD293 pKa = 3.16VFVSSAAGIEE303 pKa = 4.26LFNGEE308 pKa = 4.78DD309 pKa = 3.66PVSGEE314 pKa = 3.94VGEE317 pKa = 6.43DD318 pKa = 2.93IALWDD323 pKa = 4.44AGTEE327 pKa = 4.24RR328 pKa = 11.84NGQPGVDD335 pKa = 3.62PVSAPLQAQNGGPMAGPQEE354 pKa = 4.53GVVHH358 pKa = 6.99RR359 pKa = 11.84LDD361 pKa = 5.55DD362 pKa = 4.03INDD365 pKa = 3.68GFDD368 pKa = 3.24YY369 pKa = 11.28ADD371 pKa = 3.45ASEE374 pKa = 4.48IVDD377 pKa = 3.38VTLTHH382 pKa = 7.26DD383 pKa = 3.72GDD385 pKa = 3.97GSFTVRR391 pKa = 11.84LEE393 pKa = 4.1NTSPTDD399 pKa = 3.37VYY401 pKa = 11.5GDD403 pKa = 3.62DD404 pKa = 3.88TATGGQVWITPGAWAVHH421 pKa = 5.93TGSNPIFEE429 pKa = 4.85RR430 pKa = 11.84GEE432 pKa = 3.77PASIGLEE439 pKa = 3.91ALAEE443 pKa = 4.44AGPPTGFEE451 pKa = 4.15GQPGLVDD458 pKa = 4.23EE459 pKa = 5.58LPEE462 pKa = 5.43ADD464 pKa = 3.37NVVDD468 pKa = 3.9VGAYY472 pKa = 7.68TPSNTVTDD480 pKa = 4.59PNDD483 pKa = 3.24PMGAVPGAPPIAPGGAFEE501 pKa = 4.61FTVEE505 pKa = 4.0ADD507 pKa = 3.25PSEE510 pKa = 4.26RR511 pKa = 11.84LSFASMYY518 pKa = 10.48VPSNDD523 pKa = 2.84VFIAPEE529 pKa = 3.92PAIEE533 pKa = 5.19LYY535 pKa = 10.29TDD537 pKa = 3.61DD538 pKa = 4.82GEE540 pKa = 4.33PVEE543 pKa = 4.97GGVTEE548 pKa = 5.86SVGLWDD554 pKa = 5.93AGTEE558 pKa = 4.2LNGRR562 pKa = 11.84PGVDD566 pKa = 3.25PASAPLQAQNGGPTVGPQEE585 pKa = 4.46GVVHH589 pKa = 6.95RR590 pKa = 11.84LDD592 pKa = 6.01DD593 pKa = 3.97IDD595 pKa = 6.33DD596 pKa = 4.33GFDD599 pKa = 3.27YY600 pKa = 11.38ADD602 pKa = 3.47ASEE605 pKa = 4.39IVSVTIEE612 pKa = 3.96PQQ614 pKa = 2.82
Molecular weight: 62.98 kDa
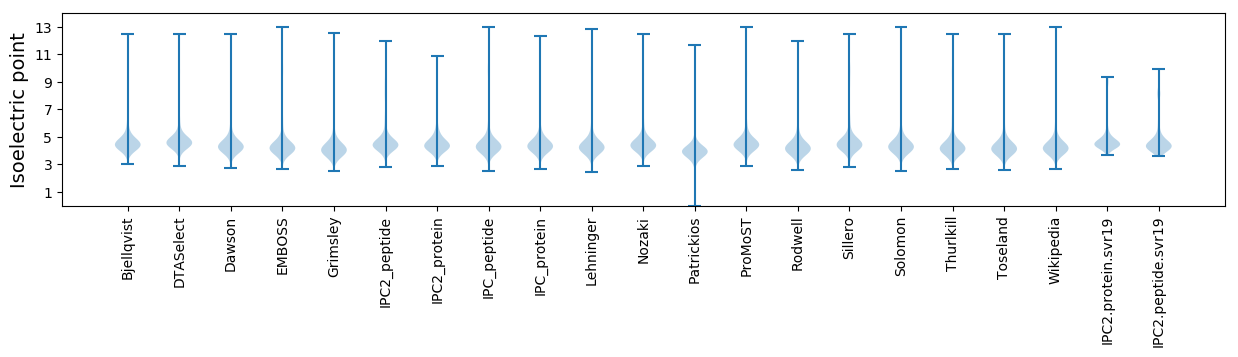
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V4Y017|V4Y017_9ARCH ABC-type dipeptide/oligopeptide/nickel transport system permease component OS=uncultured archaeon A07HR67 OX=1412871 GN=A07HR67_01204 PE=3 SV=1
MM1 pKa = 7.48ARR3 pKa = 11.84FDD5 pKa = 3.4AAEE8 pKa = 3.94RR9 pKa = 11.84RR10 pKa = 11.84NLDD13 pKa = 2.96RR14 pKa = 11.84QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NAPDD27 pKa = 3.69ADD29 pKa = 3.85RR30 pKa = 11.84CRR32 pKa = 11.84KK33 pKa = 9.62CGYY36 pKa = 9.02TNLRR40 pKa = 11.84PKK42 pKa = 10.52AKK44 pKa = 9.72EE45 pKa = 3.61RR46 pKa = 11.84RR47 pKa = 11.84ATT49 pKa = 3.4
MM1 pKa = 7.48ARR3 pKa = 11.84FDD5 pKa = 3.4AAEE8 pKa = 3.94RR9 pKa = 11.84RR10 pKa = 11.84NLDD13 pKa = 2.96RR14 pKa = 11.84QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NAPDD27 pKa = 3.69ADD29 pKa = 3.85RR30 pKa = 11.84CRR32 pKa = 11.84KK33 pKa = 9.62CGYY36 pKa = 9.02TNLRR40 pKa = 11.84PKK42 pKa = 10.52AKK44 pKa = 9.72EE45 pKa = 3.61RR46 pKa = 11.84RR47 pKa = 11.84ATT49 pKa = 3.4
Molecular weight: 5.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
769015 |
23 |
1860 |
266.4 |
28.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.351 ± 0.088 | 0.701 ± 0.015 |
8.576 ± 0.064 | 7.803 ± 0.053 |
3.17 ± 0.031 | 8.895 ± 0.042 |
1.915 ± 0.023 | 3.706 ± 0.038 |
1.472 ± 0.025 | 8.464 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.658 ± 0.019 | 2.086 ± 0.025 |
4.956 ± 0.03 | 2.01 ± 0.025 |
7.295 ± 0.054 | 5.182 ± 0.037 |
6.583 ± 0.036 | 9.576 ± 0.053 |
1.04 ± 0.019 | 2.455 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
