
Methanocella arvoryzae (strain DSM 22066 / NBRC 105507 / MRE50)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanocellales; Methanocellaceae; Methanocella; Methanocella arvoryzae
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
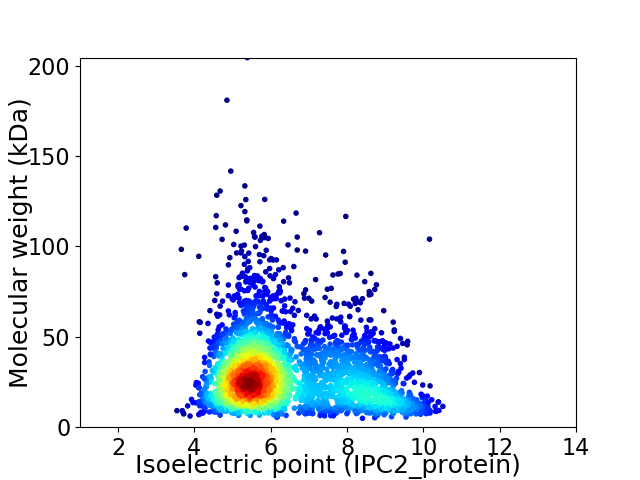
Virtual 2D-PAGE plot for 3071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0W6E3|Q0W6E3_METAR Glutamine synthetase OS=Methanocella arvoryzae (strain DSM 22066 / NBRC 105507 / MRE50) OX=351160 GN=glnA-1 PE=3 SV=1
MM1 pKa = 7.29IAASALFAAPALADD15 pKa = 4.06FPKK18 pKa = 10.82SSDD21 pKa = 3.21IQFSQSNYY29 pKa = 10.56DD30 pKa = 3.43VYY32 pKa = 11.53EE33 pKa = 4.0LANGNDD39 pKa = 3.19VTVRR43 pKa = 11.84ITVTINDD50 pKa = 4.58LDD52 pKa = 4.87LDD54 pKa = 4.0DD55 pKa = 5.04DD56 pKa = 3.66EE57 pKa = 6.23RR58 pKa = 11.84YY59 pKa = 9.45RR60 pKa = 11.84VHH62 pKa = 7.22INTDD66 pKa = 2.48QRR68 pKa = 11.84TALRR72 pKa = 11.84DD73 pKa = 3.24EE74 pKa = 4.44DD75 pKa = 5.3RR76 pKa = 11.84YY77 pKa = 10.35GQRR80 pKa = 11.84DD81 pKa = 3.67YY82 pKa = 11.73VPYY85 pKa = 9.56WEE87 pKa = 4.42HH88 pKa = 7.54DD89 pKa = 3.44YY90 pKa = 11.05RR91 pKa = 11.84YY92 pKa = 10.68LEE94 pKa = 4.19FTSADD99 pKa = 3.3NGAIAQKK106 pKa = 10.5HH107 pKa = 5.13FDD109 pKa = 3.77VTVHH113 pKa = 6.41SNTVDD118 pKa = 3.62DD119 pKa = 4.72AATEE123 pKa = 4.2YY124 pKa = 10.7FVVEE128 pKa = 3.69MRR130 pKa = 11.84GYY132 pKa = 8.32WKK134 pKa = 10.47GYY136 pKa = 6.81KK137 pKa = 8.77TLIGWIYY144 pKa = 10.08PDD146 pKa = 3.51YY147 pKa = 11.52DD148 pKa = 3.35SFKK151 pKa = 10.68NIKK154 pKa = 9.52RR155 pKa = 11.84SNVTIYY161 pKa = 10.89DD162 pKa = 3.94ADD164 pKa = 3.74TTAPITTATASGTSGANGWYY184 pKa = 8.97TSIVQVTLTASDD196 pKa = 3.89EE197 pKa = 4.37DD198 pKa = 3.84SGVANTYY205 pKa = 7.74YY206 pKa = 10.47TIDD209 pKa = 3.53GGAQQTYY216 pKa = 9.6SAPFTVTDD224 pKa = 3.96GTHH227 pKa = 7.09TIQYY231 pKa = 9.01WSVDD235 pKa = 3.07KK236 pKa = 11.41AGNTEE241 pKa = 4.05TAQTLNLNVDD251 pKa = 3.92TKK253 pKa = 11.52APVITSDD260 pKa = 3.55RR261 pKa = 11.84TPANANGWNNGDD273 pKa = 3.52VTVSFTATDD282 pKa = 3.91DD283 pKa = 3.73GSGISGSASYY293 pKa = 10.78SQTLTGEE300 pKa = 4.5GSSQSVEE307 pKa = 3.73WSVTDD312 pKa = 4.7LAGNTANATVSNINIDD328 pKa = 3.28KK329 pKa = 7.95TAPSITGAVDD339 pKa = 3.43RR340 pKa = 11.84DD341 pKa = 3.6ANANGWYY348 pKa = 9.92NADD351 pKa = 3.38VTVTFTAGADD361 pKa = 3.5LSGIASITDD370 pKa = 3.34PVVLGEE376 pKa = 4.24GAGQSVTGTVVDD388 pKa = 4.23LAGNSASTTVSGINVDD404 pKa = 2.96KK405 pKa = 9.92TAPVLVSSGPTTSPNINGWYY425 pKa = 10.22NGDD428 pKa = 3.39VTVNFVGSDD437 pKa = 3.37ALSGISGSTSADD449 pKa = 3.0VVVSTEE455 pKa = 4.01GADD458 pKa = 3.5EE459 pKa = 4.15VASYY463 pKa = 10.06TFTDD467 pKa = 3.37KK468 pKa = 11.26AGNSATFYY476 pKa = 9.65VTGIKK481 pKa = 9.82LDD483 pKa = 3.37KK484 pKa = 8.64TAPVITTARR493 pKa = 11.84TPANANGWNNGPVTVSFTATDD514 pKa = 3.4AGSGIDD520 pKa = 3.86GDD522 pKa = 3.85ATYY525 pKa = 10.72EE526 pKa = 3.96EE527 pKa = 5.05TVAGDD532 pKa = 4.21GEE534 pKa = 4.22DD535 pKa = 4.75QEE537 pKa = 5.7ASWSVSDD544 pKa = 4.72LAGNSASATVGGINIDD560 pKa = 3.56TAAPTTTDD568 pKa = 4.17DD569 pKa = 4.11LTGTLGNDD577 pKa = 2.81GWFLTDD583 pKa = 2.77VHH585 pKa = 5.85VTLSATDD592 pKa = 3.77GAAFQAVSGSGLLLLAAGSSDD613 pKa = 3.42EE614 pKa = 4.23GSSGIAEE621 pKa = 3.79IWYY624 pKa = 8.46TLDD627 pKa = 2.89GSAPIAYY634 pKa = 8.44PDD636 pKa = 3.51GGFDD640 pKa = 3.32VTGNGQHH647 pKa = 6.81VITFWSVDD655 pKa = 3.07AAGNAEE661 pKa = 4.59EE662 pKa = 4.85PNTDD666 pKa = 2.92NFKK669 pKa = 10.19IDD671 pKa = 3.17KK672 pKa = 9.05DD673 pKa = 3.94APVTTFSVTGAEE685 pKa = 4.32VTLDD689 pKa = 3.32AADD692 pKa = 4.71NIDD695 pKa = 3.62GSGVAATYY703 pKa = 7.58YY704 pKa = 9.65TLNGGLTQTYY714 pKa = 9.35IGSFPLPDD722 pKa = 3.21GTYY725 pKa = 9.99TIEE728 pKa = 4.16FWSEE732 pKa = 3.56DD733 pKa = 3.23VAGNVEE739 pKa = 4.04LHH741 pKa = 6.61GDD743 pKa = 3.87GLNTEE748 pKa = 4.39TFTVEE753 pKa = 4.12TVQSYY758 pKa = 8.46TIHH761 pKa = 7.49LLTGWNLVSSPLIIEE776 pKa = 4.34PMRR779 pKa = 11.84ASDD782 pKa = 3.44IVGNGITMVAKK793 pKa = 9.99YY794 pKa = 10.63NKK796 pKa = 8.83DD797 pKa = 2.97TGEE800 pKa = 3.82FTIYY804 pKa = 10.81SMEE807 pKa = 4.1INQPGDD813 pKa = 3.31PEE815 pKa = 4.84DD816 pKa = 3.94FVVTNDD822 pKa = 2.81VGYY825 pKa = 10.97YY826 pKa = 9.63FAASQDD832 pKa = 2.92MDD834 pKa = 3.75YY835 pKa = 10.77TFVGVYY841 pKa = 10.53APVPYY846 pKa = 10.49SIDD849 pKa = 3.45VPGDD853 pKa = 3.63NRR855 pKa = 11.84WDD857 pKa = 3.35IYY859 pKa = 11.32GWTSLQSSTAYY870 pKa = 9.7EE871 pKa = 4.37VASLMDD877 pKa = 4.42GMQMIAVYY885 pKa = 10.63NHH887 pKa = 6.53DD888 pKa = 3.7TGGFTIFSEE897 pKa = 4.92EE898 pKa = 4.04INTVQDD904 pKa = 3.15AEE906 pKa = 4.31NFVMGPGVGYY916 pKa = 9.98FITSDD921 pKa = 3.25TTTTLAYY928 pKa = 8.36EE929 pKa = 4.5VPVV932 pKa = 3.52
MM1 pKa = 7.29IAASALFAAPALADD15 pKa = 4.06FPKK18 pKa = 10.82SSDD21 pKa = 3.21IQFSQSNYY29 pKa = 10.56DD30 pKa = 3.43VYY32 pKa = 11.53EE33 pKa = 4.0LANGNDD39 pKa = 3.19VTVRR43 pKa = 11.84ITVTINDD50 pKa = 4.58LDD52 pKa = 4.87LDD54 pKa = 4.0DD55 pKa = 5.04DD56 pKa = 3.66EE57 pKa = 6.23RR58 pKa = 11.84YY59 pKa = 9.45RR60 pKa = 11.84VHH62 pKa = 7.22INTDD66 pKa = 2.48QRR68 pKa = 11.84TALRR72 pKa = 11.84DD73 pKa = 3.24EE74 pKa = 4.44DD75 pKa = 5.3RR76 pKa = 11.84YY77 pKa = 10.35GQRR80 pKa = 11.84DD81 pKa = 3.67YY82 pKa = 11.73VPYY85 pKa = 9.56WEE87 pKa = 4.42HH88 pKa = 7.54DD89 pKa = 3.44YY90 pKa = 11.05RR91 pKa = 11.84YY92 pKa = 10.68LEE94 pKa = 4.19FTSADD99 pKa = 3.3NGAIAQKK106 pKa = 10.5HH107 pKa = 5.13FDD109 pKa = 3.77VTVHH113 pKa = 6.41SNTVDD118 pKa = 3.62DD119 pKa = 4.72AATEE123 pKa = 4.2YY124 pKa = 10.7FVVEE128 pKa = 3.69MRR130 pKa = 11.84GYY132 pKa = 8.32WKK134 pKa = 10.47GYY136 pKa = 6.81KK137 pKa = 8.77TLIGWIYY144 pKa = 10.08PDD146 pKa = 3.51YY147 pKa = 11.52DD148 pKa = 3.35SFKK151 pKa = 10.68NIKK154 pKa = 9.52RR155 pKa = 11.84SNVTIYY161 pKa = 10.89DD162 pKa = 3.94ADD164 pKa = 3.74TTAPITTATASGTSGANGWYY184 pKa = 8.97TSIVQVTLTASDD196 pKa = 3.89EE197 pKa = 4.37DD198 pKa = 3.84SGVANTYY205 pKa = 7.74YY206 pKa = 10.47TIDD209 pKa = 3.53GGAQQTYY216 pKa = 9.6SAPFTVTDD224 pKa = 3.96GTHH227 pKa = 7.09TIQYY231 pKa = 9.01WSVDD235 pKa = 3.07KK236 pKa = 11.41AGNTEE241 pKa = 4.05TAQTLNLNVDD251 pKa = 3.92TKK253 pKa = 11.52APVITSDD260 pKa = 3.55RR261 pKa = 11.84TPANANGWNNGDD273 pKa = 3.52VTVSFTATDD282 pKa = 3.91DD283 pKa = 3.73GSGISGSASYY293 pKa = 10.78SQTLTGEE300 pKa = 4.5GSSQSVEE307 pKa = 3.73WSVTDD312 pKa = 4.7LAGNTANATVSNINIDD328 pKa = 3.28KK329 pKa = 7.95TAPSITGAVDD339 pKa = 3.43RR340 pKa = 11.84DD341 pKa = 3.6ANANGWYY348 pKa = 9.92NADD351 pKa = 3.38VTVTFTAGADD361 pKa = 3.5LSGIASITDD370 pKa = 3.34PVVLGEE376 pKa = 4.24GAGQSVTGTVVDD388 pKa = 4.23LAGNSASTTVSGINVDD404 pKa = 2.96KK405 pKa = 9.92TAPVLVSSGPTTSPNINGWYY425 pKa = 10.22NGDD428 pKa = 3.39VTVNFVGSDD437 pKa = 3.37ALSGISGSTSADD449 pKa = 3.0VVVSTEE455 pKa = 4.01GADD458 pKa = 3.5EE459 pKa = 4.15VASYY463 pKa = 10.06TFTDD467 pKa = 3.37KK468 pKa = 11.26AGNSATFYY476 pKa = 9.65VTGIKK481 pKa = 9.82LDD483 pKa = 3.37KK484 pKa = 8.64TAPVITTARR493 pKa = 11.84TPANANGWNNGPVTVSFTATDD514 pKa = 3.4AGSGIDD520 pKa = 3.86GDD522 pKa = 3.85ATYY525 pKa = 10.72EE526 pKa = 3.96EE527 pKa = 5.05TVAGDD532 pKa = 4.21GEE534 pKa = 4.22DD535 pKa = 4.75QEE537 pKa = 5.7ASWSVSDD544 pKa = 4.72LAGNSASATVGGINIDD560 pKa = 3.56TAAPTTTDD568 pKa = 4.17DD569 pKa = 4.11LTGTLGNDD577 pKa = 2.81GWFLTDD583 pKa = 2.77VHH585 pKa = 5.85VTLSATDD592 pKa = 3.77GAAFQAVSGSGLLLLAAGSSDD613 pKa = 3.42EE614 pKa = 4.23GSSGIAEE621 pKa = 3.79IWYY624 pKa = 8.46TLDD627 pKa = 2.89GSAPIAYY634 pKa = 8.44PDD636 pKa = 3.51GGFDD640 pKa = 3.32VTGNGQHH647 pKa = 6.81VITFWSVDD655 pKa = 3.07AAGNAEE661 pKa = 4.59EE662 pKa = 4.85PNTDD666 pKa = 2.92NFKK669 pKa = 10.19IDD671 pKa = 3.17KK672 pKa = 9.05DD673 pKa = 3.94APVTTFSVTGAEE685 pKa = 4.32VTLDD689 pKa = 3.32AADD692 pKa = 4.71NIDD695 pKa = 3.62GSGVAATYY703 pKa = 7.58YY704 pKa = 9.65TLNGGLTQTYY714 pKa = 9.35IGSFPLPDD722 pKa = 3.21GTYY725 pKa = 9.99TIEE728 pKa = 4.16FWSEE732 pKa = 3.56DD733 pKa = 3.23VAGNVEE739 pKa = 4.04LHH741 pKa = 6.61GDD743 pKa = 3.87GLNTEE748 pKa = 4.39TFTVEE753 pKa = 4.12TVQSYY758 pKa = 8.46TIHH761 pKa = 7.49LLTGWNLVSSPLIIEE776 pKa = 4.34PMRR779 pKa = 11.84ASDD782 pKa = 3.44IVGNGITMVAKK793 pKa = 9.99YY794 pKa = 10.63NKK796 pKa = 8.83DD797 pKa = 2.97TGEE800 pKa = 3.82FTIYY804 pKa = 10.81SMEE807 pKa = 4.1INQPGDD813 pKa = 3.31PEE815 pKa = 4.84DD816 pKa = 3.94FVVTNDD822 pKa = 2.81VGYY825 pKa = 10.97YY826 pKa = 9.63FAASQDD832 pKa = 2.92MDD834 pKa = 3.75YY835 pKa = 10.77TFVGVYY841 pKa = 10.53APVPYY846 pKa = 10.49SIDD849 pKa = 3.45VPGDD853 pKa = 3.63NRR855 pKa = 11.84WDD857 pKa = 3.35IYY859 pKa = 11.32GWTSLQSSTAYY870 pKa = 9.7EE871 pKa = 4.37VASLMDD877 pKa = 4.42GMQMIAVYY885 pKa = 10.63NHH887 pKa = 6.53DD888 pKa = 3.7TGGFTIFSEE897 pKa = 4.92EE898 pKa = 4.04INTVQDD904 pKa = 3.15AEE906 pKa = 4.31NFVMGPGVGYY916 pKa = 9.98FITSDD921 pKa = 3.25TTTTLAYY928 pKa = 8.36EE929 pKa = 4.5VPVV932 pKa = 3.52
Molecular weight: 98.37 kDa
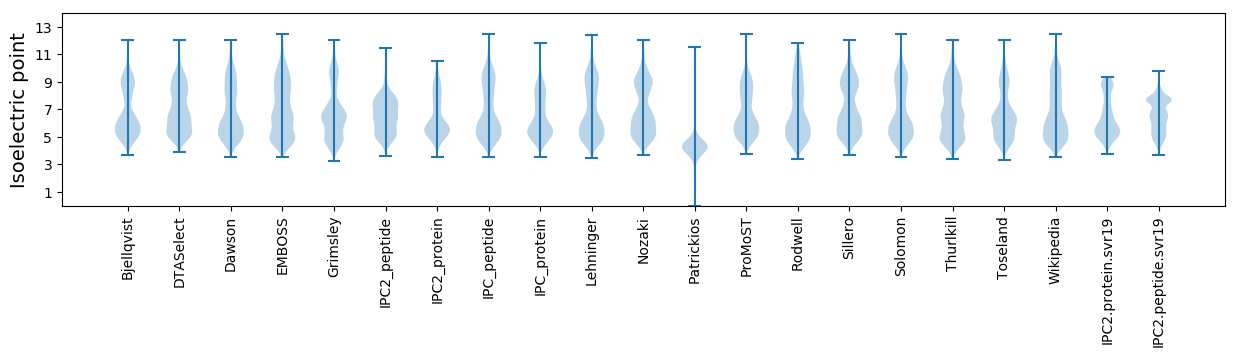
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0W584|Q0W584_METAR Uncharacterized protein OS=Methanocella arvoryzae (strain DSM 22066 / NBRC 105507 / MRE50) OX=351160 GN=RCIX1151 PE=4 SV=1
MM1 pKa = 7.4EE2 pKa = 5.84CSNKK6 pKa = 9.85CLMLACVVLALAVLTIGPAAAQLVQTGGFTRR37 pKa = 11.84TYY39 pKa = 9.0QKK41 pKa = 10.82SVAGGTFVPPQAEE54 pKa = 4.14VSEE57 pKa = 4.47TFYY60 pKa = 10.78QYY62 pKa = 11.02PGISAWGRR70 pKa = 11.84IPGRR74 pKa = 11.84WLCLRR79 pKa = 11.84LWRR82 pKa = 11.84RR83 pKa = 11.84PDD85 pKa = 3.38SVRR88 pKa = 11.84LRR90 pKa = 11.84AIRR93 pKa = 11.84RR94 pKa = 11.84LHH96 pKa = 5.56VPLRR100 pKa = 11.84NPVGKK105 pKa = 10.02KK106 pKa = 9.08ASEE109 pKa = 4.19WVSGG113 pKa = 3.63
MM1 pKa = 7.4EE2 pKa = 5.84CSNKK6 pKa = 9.85CLMLACVVLALAVLTIGPAAAQLVQTGGFTRR37 pKa = 11.84TYY39 pKa = 9.0QKK41 pKa = 10.82SVAGGTFVPPQAEE54 pKa = 4.14VSEE57 pKa = 4.47TFYY60 pKa = 10.78QYY62 pKa = 11.02PGISAWGRR70 pKa = 11.84IPGRR74 pKa = 11.84WLCLRR79 pKa = 11.84LWRR82 pKa = 11.84RR83 pKa = 11.84PDD85 pKa = 3.38SVRR88 pKa = 11.84LRR90 pKa = 11.84AIRR93 pKa = 11.84RR94 pKa = 11.84LHH96 pKa = 5.56VPLRR100 pKa = 11.84NPVGKK105 pKa = 10.02KK106 pKa = 9.08ASEE109 pKa = 4.19WVSGG113 pKa = 3.63
Molecular weight: 12.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
889879 |
44 |
1827 |
289.8 |
31.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.715 ± 0.06 | 1.297 ± 0.023 |
5.57 ± 0.037 | 6.184 ± 0.058 |
3.667 ± 0.033 | 7.808 ± 0.046 |
1.759 ± 0.019 | 7.183 ± 0.045 |
5.565 ± 0.05 | 9.24 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.967 ± 0.024 | 3.43 ± 0.034 |
4.562 ± 0.034 | 2.709 ± 0.022 |
5.446 ± 0.04 | 6.157 ± 0.038 |
5.623 ± 0.047 | 7.586 ± 0.043 |
0.943 ± 0.017 | 3.59 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
