
Kwoniella heveanensis BCC8398
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Kwoniella; Kwoniella heveanensis
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
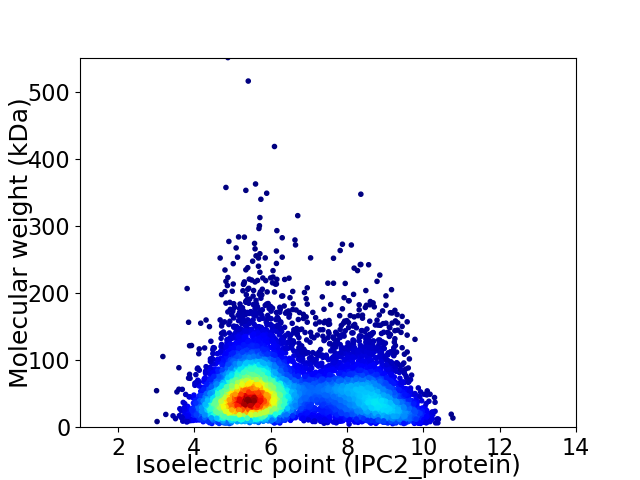
Virtual 2D-PAGE plot for 7924 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
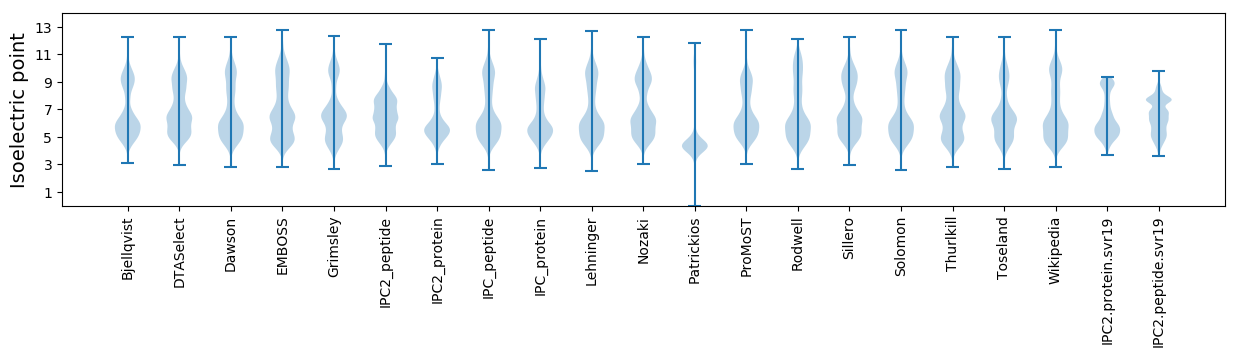
Protein with the lowest isoelectric point:
>tr|A0A1B9GJ44|A0A1B9GJ44_9TREE Cysteine proteinase 1 mitochondrial OS=Kwoniella heveanensis BCC8398 OX=1296120 GN=I316_07361 PE=3 SV=1
MM1 pKa = 7.81LYY3 pKa = 10.55NRR5 pKa = 11.84LFSLLPLLPLVFSAPLPLPLLGLDD29 pKa = 4.73DD30 pKa = 5.08LAATDD35 pKa = 5.65GILDD39 pKa = 4.26PLLNTQDD46 pKa = 3.77FAVPPPSSSDD56 pKa = 3.27PSGPALGPGGLLSGLLGNDD75 pKa = 3.21NDD77 pKa = 3.9QSEE80 pKa = 4.61GLLGSLVDD88 pKa = 3.88TVTDD92 pKa = 3.06ITGPLNISLGLNTSILGTDD111 pKa = 3.87LDD113 pKa = 4.56LGLNIQLGDD122 pKa = 4.27DD123 pKa = 3.9EE124 pKa = 5.3EE125 pKa = 5.28LICGSVNGVWDD136 pKa = 3.68GTSYY140 pKa = 11.49DD141 pKa = 4.11LRR143 pKa = 11.84CVCWSVEE150 pKa = 3.8KK151 pKa = 10.98GIVIDD156 pKa = 4.06AQVGADD162 pKa = 2.97IGLGEE167 pKa = 4.87RR168 pKa = 11.84EE169 pKa = 4.17GLDD172 pKa = 3.0AFLRR176 pKa = 11.84AQIQFGGHH184 pKa = 5.46KK185 pKa = 9.67FAYY188 pKa = 8.51PAYY191 pKa = 10.21AEE193 pKa = 4.29PTCDD197 pKa = 3.05GSGGFNCPGGRR208 pKa = 11.84QANGKK213 pKa = 9.81CSAFLAAKK221 pKa = 8.72PRR223 pKa = 11.84PRR225 pKa = 11.84PIPAADD231 pKa = 3.9AGSPAPTPTPAPAAAAIPPATDD253 pKa = 3.46LQINSVPPTTISASATTTTAAAAATVNSPDD283 pKa = 3.57APQAQAQDD291 pKa = 3.62EE292 pKa = 4.83TFGIQAVPAVAAAVTQFGTTTTTSMSTEE320 pKa = 4.3TIVLPATVFVEE331 pKa = 4.73MITTTEE337 pKa = 3.61MSTIYY342 pKa = 9.97ATVTQTEE349 pKa = 4.54VSTSISTATVTSTQTSTQTQWTTATSTVSTCSTTAEE385 pKa = 4.02DD386 pKa = 3.51EE387 pKa = 4.41QINVDD392 pKa = 4.19SVAQPTTSVTGYY404 pKa = 8.5TPASSTLVTSTSSSSATPAPEE425 pKa = 3.73VTIAATPAPATGPATSGSTGGVPAPDD451 pKa = 3.9PTSTPTPAVDD461 pKa = 4.92NGDD464 pKa = 3.94GDD466 pKa = 5.55DD467 pKa = 5.46DD468 pKa = 4.36DD469 pKa = 4.83TFSGPRR475 pKa = 11.84VIDD478 pKa = 3.45LAKK481 pKa = 9.39PQSHH485 pKa = 6.78GSTTCPEE492 pKa = 3.89GQAWGSTMCCRR503 pKa = 11.84TDD505 pKa = 3.32QVEE508 pKa = 3.97VDD510 pKa = 4.63GEE512 pKa = 4.71CKK514 pKa = 10.53CHH516 pKa = 7.38DD517 pKa = 3.8GFEE520 pKa = 4.87NIVDD524 pKa = 3.62VCLRR528 pKa = 11.84LCIGDD533 pKa = 3.86RR534 pKa = 11.84LPSGEE539 pKa = 4.45CSILGLGLDD548 pKa = 4.24LGVGLGGLEE557 pKa = 4.02LL558 pKa = 4.9
MM1 pKa = 7.81LYY3 pKa = 10.55NRR5 pKa = 11.84LFSLLPLLPLVFSAPLPLPLLGLDD29 pKa = 4.73DD30 pKa = 5.08LAATDD35 pKa = 5.65GILDD39 pKa = 4.26PLLNTQDD46 pKa = 3.77FAVPPPSSSDD56 pKa = 3.27PSGPALGPGGLLSGLLGNDD75 pKa = 3.21NDD77 pKa = 3.9QSEE80 pKa = 4.61GLLGSLVDD88 pKa = 3.88TVTDD92 pKa = 3.06ITGPLNISLGLNTSILGTDD111 pKa = 3.87LDD113 pKa = 4.56LGLNIQLGDD122 pKa = 4.27DD123 pKa = 3.9EE124 pKa = 5.3EE125 pKa = 5.28LICGSVNGVWDD136 pKa = 3.68GTSYY140 pKa = 11.49DD141 pKa = 4.11LRR143 pKa = 11.84CVCWSVEE150 pKa = 3.8KK151 pKa = 10.98GIVIDD156 pKa = 4.06AQVGADD162 pKa = 2.97IGLGEE167 pKa = 4.87RR168 pKa = 11.84EE169 pKa = 4.17GLDD172 pKa = 3.0AFLRR176 pKa = 11.84AQIQFGGHH184 pKa = 5.46KK185 pKa = 9.67FAYY188 pKa = 8.51PAYY191 pKa = 10.21AEE193 pKa = 4.29PTCDD197 pKa = 3.05GSGGFNCPGGRR208 pKa = 11.84QANGKK213 pKa = 9.81CSAFLAAKK221 pKa = 8.72PRR223 pKa = 11.84PRR225 pKa = 11.84PIPAADD231 pKa = 3.9AGSPAPTPTPAPAAAAIPPATDD253 pKa = 3.46LQINSVPPTTISASATTTTAAAAATVNSPDD283 pKa = 3.57APQAQAQDD291 pKa = 3.62EE292 pKa = 4.83TFGIQAVPAVAAAVTQFGTTTTTSMSTEE320 pKa = 4.3TIVLPATVFVEE331 pKa = 4.73MITTTEE337 pKa = 3.61MSTIYY342 pKa = 9.97ATVTQTEE349 pKa = 4.54VSTSISTATVTSTQTSTQTQWTTATSTVSTCSTTAEE385 pKa = 4.02DD386 pKa = 3.51EE387 pKa = 4.41QINVDD392 pKa = 4.19SVAQPTTSVTGYY404 pKa = 8.5TPASSTLVTSTSSSSATPAPEE425 pKa = 3.73VTIAATPAPATGPATSGSTGGVPAPDD451 pKa = 3.9PTSTPTPAVDD461 pKa = 4.92NGDD464 pKa = 3.94GDD466 pKa = 5.55DD467 pKa = 5.46DD468 pKa = 4.36DD469 pKa = 4.83TFSGPRR475 pKa = 11.84VIDD478 pKa = 3.45LAKK481 pKa = 9.39PQSHH485 pKa = 6.78GSTTCPEE492 pKa = 3.89GQAWGSTMCCRR503 pKa = 11.84TDD505 pKa = 3.32QVEE508 pKa = 3.97VDD510 pKa = 4.63GEE512 pKa = 4.71CKK514 pKa = 10.53CHH516 pKa = 7.38DD517 pKa = 3.8GFEE520 pKa = 4.87NIVDD524 pKa = 3.62VCLRR528 pKa = 11.84LCIGDD533 pKa = 3.86RR534 pKa = 11.84LPSGEE539 pKa = 4.45CSILGLGLDD548 pKa = 4.24LGVGLGGLEE557 pKa = 4.02LL558 pKa = 4.9
Molecular weight: 56.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9GKX1|A0A1B9GKX1_9TREE BRCT domain-containing protein OS=Kwoniella heveanensis BCC8398 OX=1296120 GN=I316_06790 PE=4 SV=1
MM1 pKa = 7.56PASPQICEE9 pKa = 3.94NCGLPRR15 pKa = 11.84AFPPSPRR22 pKa = 11.84DD23 pKa = 3.28LPTMFEE29 pKa = 4.22EE30 pKa = 4.53DD31 pKa = 3.6FEE33 pKa = 5.16RR34 pKa = 11.84CFICEE39 pKa = 4.09KK40 pKa = 10.24ACKK43 pKa = 9.98GLYY46 pKa = 9.87CSSEE50 pKa = 3.78CRR52 pKa = 11.84LRR54 pKa = 11.84DD55 pKa = 3.24QGTPSPAVPAAKK67 pKa = 9.45MGPVKK72 pKa = 9.79ITSQLPASLSPLVRR86 pKa = 11.84PAHH89 pKa = 5.9HH90 pKa = 7.59PGRR93 pKa = 11.84SPRR96 pKa = 11.84VFAQNRR102 pKa = 11.84ASSSISSGSSSVSSSPLQSPQTNPSEE128 pKa = 4.09ADD130 pKa = 3.44SPKK133 pKa = 10.49KK134 pKa = 9.97DD135 pKa = 3.64TFDD138 pKa = 4.63LPPPAYY144 pKa = 8.39PFKK147 pKa = 11.07VGGIPTSVPMKK158 pKa = 10.23IPALIPRR165 pKa = 11.84AAVAQQPHH173 pKa = 5.98GQTPGSQGSTIYY185 pKa = 10.23PAGTSIDD192 pKa = 3.44TLRR195 pKa = 11.84FGRR198 pKa = 11.84KK199 pKa = 8.36PSVVNNVISPNALIPRR215 pKa = 11.84CACGLPANHH224 pKa = 7.6KK225 pKa = 10.21GRR227 pKa = 11.84ANSKK231 pKa = 10.36DD232 pKa = 3.45RR233 pKa = 11.84ADD235 pKa = 3.81LPEE238 pKa = 5.51SGFSRR243 pKa = 11.84LSLGPSVVNPPHH255 pKa = 6.9ASEE258 pKa = 4.41EE259 pKa = 3.97PGPRR263 pKa = 11.84SFRR266 pKa = 11.84IVSDD270 pKa = 3.41SAIPPFPGRR279 pKa = 11.84PVQPSVPTPSRR290 pKa = 11.84ASIPLSIPQSPQVNPTASLLSRR312 pKa = 11.84SRR314 pKa = 11.84SDD316 pKa = 4.84PIPSSPQVQRR326 pKa = 11.84RR327 pKa = 11.84AKK329 pKa = 9.47PIPAAPAPAPIIATNVITPSHH350 pKa = 7.11RR351 pKa = 11.84EE352 pKa = 3.71TFAVPMSPIVPAQASACRR370 pKa = 11.84PSRR373 pKa = 11.84HH374 pKa = 5.64TPNALEE380 pKa = 4.14VSMASPRR387 pKa = 11.84RR388 pKa = 11.84GRR390 pKa = 11.84SRR392 pKa = 11.84EE393 pKa = 3.94RR394 pKa = 11.84QEE396 pKa = 3.86HH397 pKa = 6.45HH398 pKa = 6.42VPQMTGNALGGPADD412 pKa = 4.23RR413 pKa = 11.84EE414 pKa = 4.23QAPSRR419 pKa = 11.84SRR421 pKa = 11.84NRR423 pKa = 11.84RR424 pKa = 11.84DD425 pKa = 2.76SRR427 pKa = 11.84RR428 pKa = 11.84RR429 pKa = 11.84SDD431 pKa = 2.95SRR433 pKa = 11.84EE434 pKa = 3.65RR435 pKa = 11.84EE436 pKa = 3.88RR437 pKa = 11.84EE438 pKa = 3.78RR439 pKa = 11.84EE440 pKa = 3.92RR441 pKa = 11.84GRR443 pKa = 11.84GRR445 pKa = 11.84AGVVVGDD452 pKa = 4.15RR453 pKa = 11.84EE454 pKa = 4.33RR455 pKa = 11.84EE456 pKa = 3.91HH457 pKa = 7.56SGQRR461 pKa = 11.84SPRR464 pKa = 11.84SPLATTNGTVDD475 pKa = 3.99PPQILPSWSRR485 pKa = 11.84RR486 pKa = 11.84ASEE489 pKa = 3.81ATADD493 pKa = 3.62MRR495 pKa = 11.84KK496 pKa = 9.48LVGDD500 pKa = 4.01GVVTPVMRR508 pKa = 11.84RR509 pKa = 11.84TSSGDD514 pKa = 2.99KK515 pKa = 10.22KK516 pKa = 10.95SPVYY520 pKa = 10.45EE521 pKa = 3.93RR522 pKa = 11.84DD523 pKa = 3.12EE524 pKa = 4.25EE525 pKa = 4.0EE526 pKa = 4.28RR527 pKa = 11.84EE528 pKa = 4.08RR529 pKa = 11.84KK530 pKa = 9.6RR531 pKa = 11.84EE532 pKa = 3.85EE533 pKa = 3.68LKK535 pKa = 10.79RR536 pKa = 11.84ASKK539 pKa = 10.63QLGQVFGVAAGG550 pKa = 3.38
MM1 pKa = 7.56PASPQICEE9 pKa = 3.94NCGLPRR15 pKa = 11.84AFPPSPRR22 pKa = 11.84DD23 pKa = 3.28LPTMFEE29 pKa = 4.22EE30 pKa = 4.53DD31 pKa = 3.6FEE33 pKa = 5.16RR34 pKa = 11.84CFICEE39 pKa = 4.09KK40 pKa = 10.24ACKK43 pKa = 9.98GLYY46 pKa = 9.87CSSEE50 pKa = 3.78CRR52 pKa = 11.84LRR54 pKa = 11.84DD55 pKa = 3.24QGTPSPAVPAAKK67 pKa = 9.45MGPVKK72 pKa = 9.79ITSQLPASLSPLVRR86 pKa = 11.84PAHH89 pKa = 5.9HH90 pKa = 7.59PGRR93 pKa = 11.84SPRR96 pKa = 11.84VFAQNRR102 pKa = 11.84ASSSISSGSSSVSSSPLQSPQTNPSEE128 pKa = 4.09ADD130 pKa = 3.44SPKK133 pKa = 10.49KK134 pKa = 9.97DD135 pKa = 3.64TFDD138 pKa = 4.63LPPPAYY144 pKa = 8.39PFKK147 pKa = 11.07VGGIPTSVPMKK158 pKa = 10.23IPALIPRR165 pKa = 11.84AAVAQQPHH173 pKa = 5.98GQTPGSQGSTIYY185 pKa = 10.23PAGTSIDD192 pKa = 3.44TLRR195 pKa = 11.84FGRR198 pKa = 11.84KK199 pKa = 8.36PSVVNNVISPNALIPRR215 pKa = 11.84CACGLPANHH224 pKa = 7.6KK225 pKa = 10.21GRR227 pKa = 11.84ANSKK231 pKa = 10.36DD232 pKa = 3.45RR233 pKa = 11.84ADD235 pKa = 3.81LPEE238 pKa = 5.51SGFSRR243 pKa = 11.84LSLGPSVVNPPHH255 pKa = 6.9ASEE258 pKa = 4.41EE259 pKa = 3.97PGPRR263 pKa = 11.84SFRR266 pKa = 11.84IVSDD270 pKa = 3.41SAIPPFPGRR279 pKa = 11.84PVQPSVPTPSRR290 pKa = 11.84ASIPLSIPQSPQVNPTASLLSRR312 pKa = 11.84SRR314 pKa = 11.84SDD316 pKa = 4.84PIPSSPQVQRR326 pKa = 11.84RR327 pKa = 11.84AKK329 pKa = 9.47PIPAAPAPAPIIATNVITPSHH350 pKa = 7.11RR351 pKa = 11.84EE352 pKa = 3.71TFAVPMSPIVPAQASACRR370 pKa = 11.84PSRR373 pKa = 11.84HH374 pKa = 5.64TPNALEE380 pKa = 4.14VSMASPRR387 pKa = 11.84RR388 pKa = 11.84GRR390 pKa = 11.84SRR392 pKa = 11.84EE393 pKa = 3.94RR394 pKa = 11.84QEE396 pKa = 3.86HH397 pKa = 6.45HH398 pKa = 6.42VPQMTGNALGGPADD412 pKa = 4.23RR413 pKa = 11.84EE414 pKa = 4.23QAPSRR419 pKa = 11.84SRR421 pKa = 11.84NRR423 pKa = 11.84RR424 pKa = 11.84DD425 pKa = 2.76SRR427 pKa = 11.84RR428 pKa = 11.84RR429 pKa = 11.84SDD431 pKa = 2.95SRR433 pKa = 11.84EE434 pKa = 3.65RR435 pKa = 11.84EE436 pKa = 3.88RR437 pKa = 11.84EE438 pKa = 3.78RR439 pKa = 11.84EE440 pKa = 3.92RR441 pKa = 11.84GRR443 pKa = 11.84GRR445 pKa = 11.84AGVVVGDD452 pKa = 4.15RR453 pKa = 11.84EE454 pKa = 4.33RR455 pKa = 11.84EE456 pKa = 3.91HH457 pKa = 7.56SGQRR461 pKa = 11.84SPRR464 pKa = 11.84SPLATTNGTVDD475 pKa = 3.99PPQILPSWSRR485 pKa = 11.84RR486 pKa = 11.84ASEE489 pKa = 3.81ATADD493 pKa = 3.62MRR495 pKa = 11.84KK496 pKa = 9.48LVGDD500 pKa = 4.01GVVTPVMRR508 pKa = 11.84RR509 pKa = 11.84TSSGDD514 pKa = 2.99KK515 pKa = 10.22KK516 pKa = 10.95SPVYY520 pKa = 10.45EE521 pKa = 3.93RR522 pKa = 11.84DD523 pKa = 3.12EE524 pKa = 4.25EE525 pKa = 4.0EE526 pKa = 4.28RR527 pKa = 11.84EE528 pKa = 4.08RR529 pKa = 11.84KK530 pKa = 9.6RR531 pKa = 11.84EE532 pKa = 3.85EE533 pKa = 3.68LKK535 pKa = 10.79RR536 pKa = 11.84ASKK539 pKa = 10.63QLGQVFGVAAGG550 pKa = 3.38
Molecular weight: 59.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4255186 |
44 |
5007 |
537.0 |
58.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.998 ± 0.028 | 0.937 ± 0.009 |
5.611 ± 0.019 | 6.203 ± 0.026 |
3.188 ± 0.016 | 7.705 ± 0.026 |
2.369 ± 0.013 | 4.544 ± 0.016 |
4.738 ± 0.022 | 8.54 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.019 ± 0.011 | 3.325 ± 0.014 |
6.75 ± 0.033 | 3.849 ± 0.022 |
6.015 ± 0.023 | 9.498 ± 0.036 |
6.046 ± 0.019 | 5.907 ± 0.018 |
1.316 ± 0.01 | 2.442 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
