
Strongyloides ratti (Parasitic roundworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Tylenchina; Panagrolaimomorpha; Strongyloidoidea; Strongyloididae; Strongyloides
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
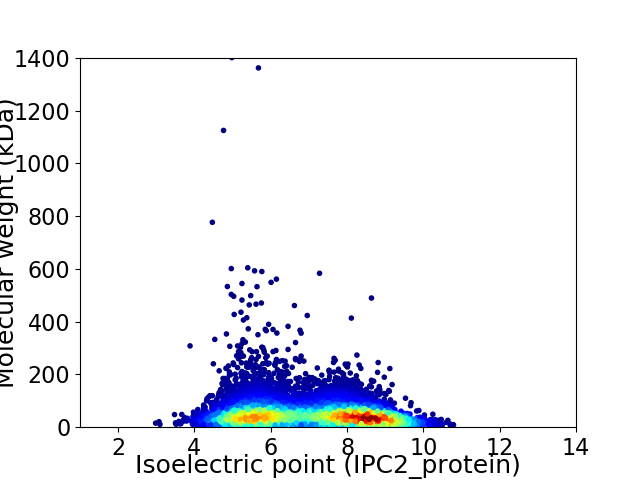
Virtual 2D-PAGE plot for 9956 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A090L4J0|A0A090L4J0_STRRB Thrombospondin type 1 repeat-containing protein OS=Strongyloides ratti OX=34506 GN=SRAE_1000068700 PE=4 SV=1
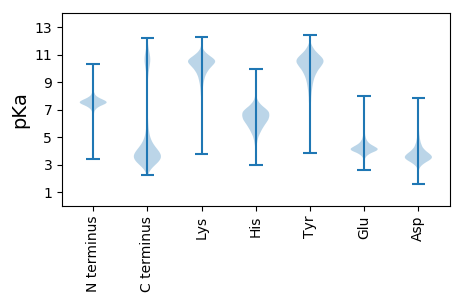
MM1 pKa = 7.82LYY3 pKa = 10.49NLSFDD8 pKa = 3.91VLSDD12 pKa = 3.21KK13 pKa = 10.54STSQEE18 pKa = 3.82NNVLLKK24 pKa = 10.93LPNNDD29 pKa = 2.25VTTQNIPTEE38 pKa = 4.4SEE40 pKa = 4.16EE41 pKa = 4.13EE42 pKa = 3.94MSDD45 pKa = 3.14PFVFTTSIPSVSPQPPGIDD64 pKa = 3.33LSFDD68 pKa = 3.63NNLEE72 pKa = 4.14TTTLDD77 pKa = 3.47SLIANSRR84 pKa = 11.84TTNFIEE90 pKa = 5.33LSTLKK95 pKa = 10.77DD96 pKa = 3.63DD97 pKa = 6.6FNLEE101 pKa = 4.27DD102 pKa = 4.36NDD104 pKa = 4.83TIEE107 pKa = 5.53DD108 pKa = 3.79DD109 pKa = 3.94QINITVAPLNSSKK122 pKa = 10.47QVSLSNDD129 pKa = 3.01FDD131 pKa = 4.23NKK133 pKa = 9.8TSEE136 pKa = 4.09GVNNFSNFDD145 pKa = 3.51INDD148 pKa = 3.42QYY150 pKa = 12.05VSLISKK156 pKa = 8.86EE157 pKa = 4.12AFITTTPANEE167 pKa = 4.41LNDD170 pKa = 3.73NDD172 pKa = 4.67EE173 pKa = 5.43LDD175 pKa = 3.99NLDD178 pKa = 4.45DD179 pKa = 5.75NEE181 pKa = 4.35IFLNTSSTTTILPKK195 pKa = 9.75MVNEE199 pKa = 4.82NIPLTTEE206 pKa = 4.25VYY208 pKa = 7.32PTNITSTSEE217 pKa = 3.86EE218 pKa = 3.91VHH220 pKa = 6.65LNNSDD225 pKa = 3.28KK226 pKa = 11.23FIVDD230 pKa = 4.28EE231 pKa = 4.3YY232 pKa = 11.03NGNSIEE238 pKa = 4.14NKK240 pKa = 7.45YY241 pKa = 9.38TNDD244 pKa = 3.08IKK246 pKa = 11.01KK247 pKa = 10.0LCPEE251 pKa = 4.21VEE253 pKa = 4.05ICNPGCGISIDD264 pKa = 3.78FDD266 pKa = 3.78GCQKK270 pKa = 9.05CTCLWIPNFCIEE282 pKa = 4.33TSDD285 pKa = 4.11CNGPEE290 pKa = 4.21YY291 pKa = 10.2ICEE294 pKa = 3.99YY295 pKa = 10.59GKK297 pKa = 10.4CLCDD301 pKa = 3.49INYY304 pKa = 7.56TQDD307 pKa = 3.13MEE309 pKa = 4.56RR310 pKa = 11.84SGICIPKK317 pKa = 9.99PNSLKK322 pKa = 10.96QNDD325 pKa = 3.5VV326 pKa = 3.14
MM1 pKa = 7.82LYY3 pKa = 10.49NLSFDD8 pKa = 3.91VLSDD12 pKa = 3.21KK13 pKa = 10.54STSQEE18 pKa = 3.82NNVLLKK24 pKa = 10.93LPNNDD29 pKa = 2.25VTTQNIPTEE38 pKa = 4.4SEE40 pKa = 4.16EE41 pKa = 4.13EE42 pKa = 3.94MSDD45 pKa = 3.14PFVFTTSIPSVSPQPPGIDD64 pKa = 3.33LSFDD68 pKa = 3.63NNLEE72 pKa = 4.14TTTLDD77 pKa = 3.47SLIANSRR84 pKa = 11.84TTNFIEE90 pKa = 5.33LSTLKK95 pKa = 10.77DD96 pKa = 3.63DD97 pKa = 6.6FNLEE101 pKa = 4.27DD102 pKa = 4.36NDD104 pKa = 4.83TIEE107 pKa = 5.53DD108 pKa = 3.79DD109 pKa = 3.94QINITVAPLNSSKK122 pKa = 10.47QVSLSNDD129 pKa = 3.01FDD131 pKa = 4.23NKK133 pKa = 9.8TSEE136 pKa = 4.09GVNNFSNFDD145 pKa = 3.51INDD148 pKa = 3.42QYY150 pKa = 12.05VSLISKK156 pKa = 8.86EE157 pKa = 4.12AFITTTPANEE167 pKa = 4.41LNDD170 pKa = 3.73NDD172 pKa = 4.67EE173 pKa = 5.43LDD175 pKa = 3.99NLDD178 pKa = 4.45DD179 pKa = 5.75NEE181 pKa = 4.35IFLNTSSTTTILPKK195 pKa = 9.75MVNEE199 pKa = 4.82NIPLTTEE206 pKa = 4.25VYY208 pKa = 7.32PTNITSTSEE217 pKa = 3.86EE218 pKa = 3.91VHH220 pKa = 6.65LNNSDD225 pKa = 3.28KK226 pKa = 11.23FIVDD230 pKa = 4.28EE231 pKa = 4.3YY232 pKa = 11.03NGNSIEE238 pKa = 4.14NKK240 pKa = 7.45YY241 pKa = 9.38TNDD244 pKa = 3.08IKK246 pKa = 11.01KK247 pKa = 10.0LCPEE251 pKa = 4.21VEE253 pKa = 4.05ICNPGCGISIDD264 pKa = 3.78FDD266 pKa = 3.78GCQKK270 pKa = 9.05CTCLWIPNFCIEE282 pKa = 4.33TSDD285 pKa = 4.11CNGPEE290 pKa = 4.21YY291 pKa = 10.2ICEE294 pKa = 3.99YY295 pKa = 10.59GKK297 pKa = 10.4CLCDD301 pKa = 3.49INYY304 pKa = 7.56TQDD307 pKa = 3.13MEE309 pKa = 4.56RR310 pKa = 11.84SGICIPKK317 pKa = 9.99PNSLKK322 pKa = 10.96QNDD325 pKa = 3.5VV326 pKa = 3.14
Molecular weight: 36.44 kDa
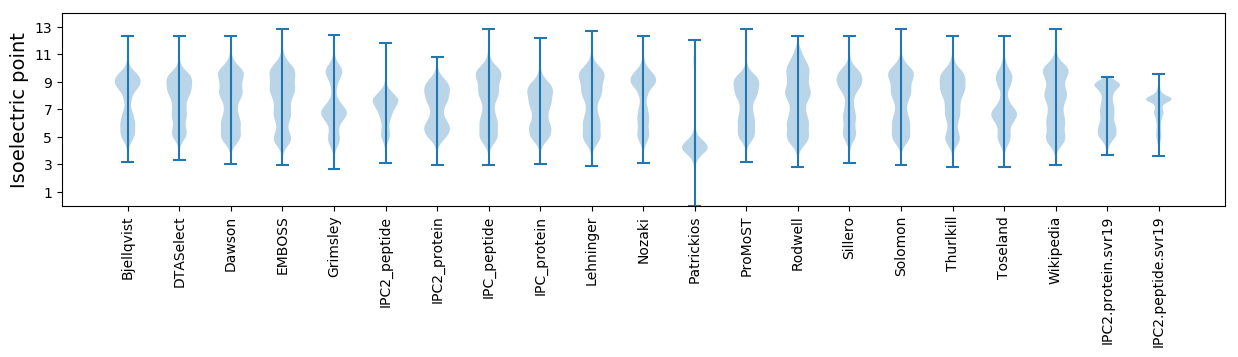
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A090LSW4|A0A090LSW4_STRRB Poly-glutamine tract binding protein 1 OS=Strongyloides ratti OX=34506 GN=SRAE_X000061900 PE=4 SV=1
MM1 pKa = 7.21FKK3 pKa = 10.42IMNLLNFNFFVIFFGIFIFTINGCQLKK30 pKa = 10.46YY31 pKa = 10.88SGIFGRR37 pKa = 11.84KK38 pKa = 6.69QCSLVINLNFDD49 pKa = 3.78PFFKK53 pKa = 10.43MSAFCLHH60 pKa = 6.18SAVYY64 pKa = 10.46QNLLRR69 pKa = 11.84MNKK72 pKa = 8.58QRR74 pKa = 11.84MGMNPKK80 pKa = 10.39LNAKK84 pKa = 10.04LLMDD88 pKa = 3.51MTMYY92 pKa = 11.2ANNNIGPKK100 pKa = 10.31NIMQLCTQCGTQCGMQTKK118 pKa = 9.8IKK120 pKa = 10.52FIGDD124 pKa = 4.49PIQGQGSLRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84GSLTLSQRR143 pKa = 11.84RR144 pKa = 11.84KK145 pKa = 10.18NVFDD149 pKa = 3.89NAIHH153 pKa = 7.77DD154 pKa = 4.62DD155 pKa = 4.04ADD157 pKa = 3.77DD158 pKa = 5.06PIYY161 pKa = 10.78ATVNRR166 pKa = 11.84NRR168 pKa = 11.84FRR170 pKa = 11.84GSINRR175 pKa = 11.84GNSFNRR181 pKa = 11.84GGSFHH186 pKa = 7.31SNGGSFRR193 pKa = 11.84GGGGSFRR200 pKa = 11.84NGGGFNGRR208 pKa = 11.84GSLHH212 pKa = 6.73RR213 pKa = 11.84SNSVEE218 pKa = 3.57QRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84AVFANEE228 pKa = 4.18RR229 pKa = 11.84LDD231 pKa = 3.6SDD233 pKa = 3.94EE234 pKa = 4.02
MM1 pKa = 7.21FKK3 pKa = 10.42IMNLLNFNFFVIFFGIFIFTINGCQLKK30 pKa = 10.46YY31 pKa = 10.88SGIFGRR37 pKa = 11.84KK38 pKa = 6.69QCSLVINLNFDD49 pKa = 3.78PFFKK53 pKa = 10.43MSAFCLHH60 pKa = 6.18SAVYY64 pKa = 10.46QNLLRR69 pKa = 11.84MNKK72 pKa = 8.58QRR74 pKa = 11.84MGMNPKK80 pKa = 10.39LNAKK84 pKa = 10.04LLMDD88 pKa = 3.51MTMYY92 pKa = 11.2ANNNIGPKK100 pKa = 10.31NIMQLCTQCGTQCGMQTKK118 pKa = 9.8IKK120 pKa = 10.52FIGDD124 pKa = 4.49PIQGQGSLRR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84GSLTLSQRR143 pKa = 11.84RR144 pKa = 11.84KK145 pKa = 10.18NVFDD149 pKa = 3.89NAIHH153 pKa = 7.77DD154 pKa = 4.62DD155 pKa = 4.04ADD157 pKa = 3.77DD158 pKa = 5.06PIYY161 pKa = 10.78ATVNRR166 pKa = 11.84NRR168 pKa = 11.84FRR170 pKa = 11.84GSINRR175 pKa = 11.84GNSFNRR181 pKa = 11.84GGSFHH186 pKa = 7.31SNGGSFRR193 pKa = 11.84GGGGSFRR200 pKa = 11.84NGGGFNGRR208 pKa = 11.84GSLHH212 pKa = 6.73RR213 pKa = 11.84SNSVEE218 pKa = 3.57QRR220 pKa = 11.84RR221 pKa = 11.84RR222 pKa = 11.84AVFANEE228 pKa = 4.18RR229 pKa = 11.84LDD231 pKa = 3.6SDD233 pKa = 3.94EE234 pKa = 4.02
Molecular weight: 26.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4680237 |
14 |
12256 |
470.1 |
53.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.902 ± 0.022 | 2.063 ± 0.019 |
5.51 ± 0.02 | 6.64 ± 0.032 |
4.906 ± 0.026 | 4.755 ± 0.031 |
1.953 ± 0.009 | 8.57 ± 0.029 |
8.621 ± 0.031 | 8.628 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.011 | 7.619 ± 0.034 |
3.835 ± 0.026 | 3.372 ± 0.017 |
3.831 ± 0.025 | 8.016 ± 0.029 |
5.557 ± 0.039 | 5.051 ± 0.019 |
0.888 ± 0.008 | 4.063 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
