
Torque teno virus
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
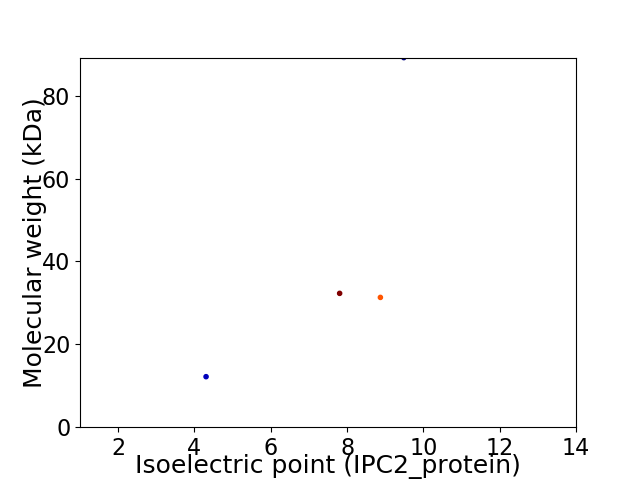
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
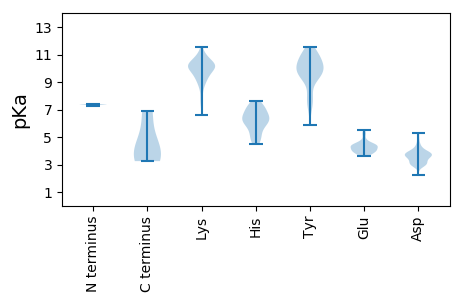
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q8V7H8|Q8V7H8_9VIRU Capsid protein OS=Torque teno virus OX=68887 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.39PWSLPRR7 pKa = 11.84HH8 pKa = 5.86NIRR11 pKa = 11.84TRR13 pKa = 11.84EE14 pKa = 4.01DD15 pKa = 3.01LWVQSILYY23 pKa = 10.14SHH25 pKa = 7.23DD26 pKa = 3.69TFCGCDD32 pKa = 3.75NIPEE36 pKa = 4.6HH37 pKa = 6.14LTGLLGGVRR46 pKa = 11.84PAPPRR51 pKa = 11.84NPGPPTIRR59 pKa = 11.84SLPALPPAPEE69 pKa = 4.26PPEE72 pKa = 3.88EE73 pKa = 4.04PRR75 pKa = 11.84RR76 pKa = 11.84GGDD79 pKa = 3.05TDD81 pKa = 3.6GDD83 pKa = 3.75RR84 pKa = 11.84GEE86 pKa = 5.28DD87 pKa = 3.44GGDD90 pKa = 2.93AAGAYY95 pKa = 9.42EE96 pKa = 4.39PEE98 pKa = 4.29DD99 pKa = 4.0LEE101 pKa = 4.46EE102 pKa = 4.44LFAAAEE108 pKa = 3.99QDD110 pKa = 4.22DD111 pKa = 4.0MM112 pKa = 6.93
MM1 pKa = 7.39PWSLPRR7 pKa = 11.84HH8 pKa = 5.86NIRR11 pKa = 11.84TRR13 pKa = 11.84EE14 pKa = 4.01DD15 pKa = 3.01LWVQSILYY23 pKa = 10.14SHH25 pKa = 7.23DD26 pKa = 3.69TFCGCDD32 pKa = 3.75NIPEE36 pKa = 4.6HH37 pKa = 6.14LTGLLGGVRR46 pKa = 11.84PAPPRR51 pKa = 11.84NPGPPTIRR59 pKa = 11.84SLPALPPAPEE69 pKa = 4.26PPEE72 pKa = 3.88EE73 pKa = 4.04PRR75 pKa = 11.84RR76 pKa = 11.84GGDD79 pKa = 3.05TDD81 pKa = 3.6GDD83 pKa = 3.75RR84 pKa = 11.84GEE86 pKa = 5.28DD87 pKa = 3.44GGDD90 pKa = 2.93AAGAYY95 pKa = 9.42EE96 pKa = 4.39PEE98 pKa = 4.29DD99 pKa = 4.0LEE101 pKa = 4.46EE102 pKa = 4.44LFAAAEE108 pKa = 3.99QDD110 pKa = 4.22DD111 pKa = 4.0MM112 pKa = 6.93
Molecular weight: 12.19 kDa
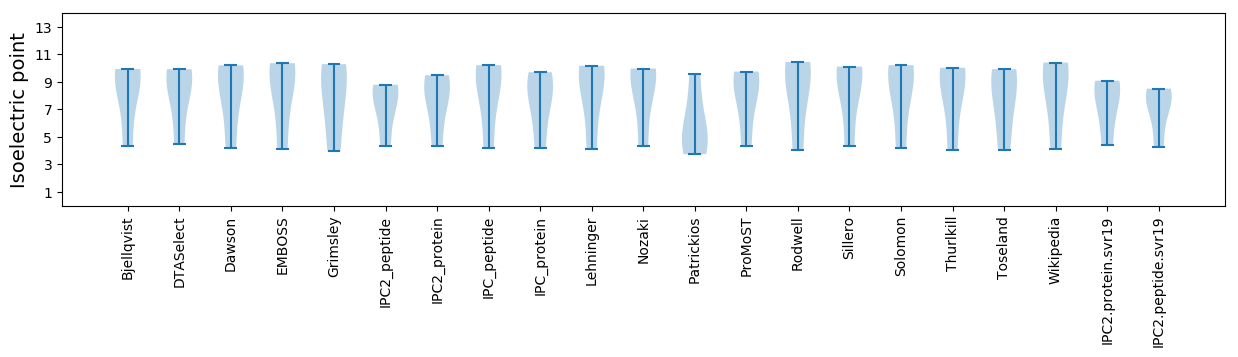
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q8V7I0|Q8V7I0_9VIRU ORF4 OS=Torque teno virus OX=68887 GN=ORF4 PE=4 SV=1
MM1 pKa = 7.39PWSLPRR7 pKa = 11.84HH8 pKa = 5.86NIRR11 pKa = 11.84TRR13 pKa = 11.84EE14 pKa = 4.01DD15 pKa = 3.01LWVQSILYY23 pKa = 10.14SHH25 pKa = 7.23DD26 pKa = 3.69TFCGCDD32 pKa = 3.75NIPEE36 pKa = 4.6HH37 pKa = 6.14LTGLLGGVRR46 pKa = 11.84PAPPRR51 pKa = 11.84NPGPPTIRR59 pKa = 11.84SLPALPPAPEE69 pKa = 4.26PPEE72 pKa = 3.88EE73 pKa = 4.04PRR75 pKa = 11.84RR76 pKa = 11.84GGDD79 pKa = 3.05TDD81 pKa = 3.6GDD83 pKa = 3.75RR84 pKa = 11.84GEE86 pKa = 5.28DD87 pKa = 3.44GGDD90 pKa = 2.93AAGAYY95 pKa = 9.42EE96 pKa = 4.39PEE98 pKa = 4.29DD99 pKa = 4.0LEE101 pKa = 4.46EE102 pKa = 4.44LFAAAEE108 pKa = 4.02QDD110 pKa = 3.82DD111 pKa = 3.88MRR113 pKa = 11.84RR114 pKa = 11.84VQKK117 pKa = 9.78TPIRR121 pKa = 11.84HH122 pKa = 5.33SPPRR126 pKa = 11.84AAIKK130 pKa = 9.45PRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LVYY138 pKa = 9.86PQTTPKK144 pKa = 10.2RR145 pKa = 11.84ARR147 pKa = 11.84RR148 pKa = 11.84DD149 pKa = 3.42HH150 pKa = 6.42LRR152 pKa = 11.84GRR154 pKa = 11.84TSTPKK159 pKa = 10.15RR160 pKa = 11.84RR161 pKa = 11.84GAKK164 pKa = 9.46RR165 pKa = 11.84KK166 pKa = 10.12APTATQSPATAPASPQTGNQTPHH189 pKa = 5.98GRR191 pKa = 11.84RR192 pKa = 11.84PPTQTGSPLEE202 pKa = 4.17PSPIILPPEE211 pKa = 5.15PIPDD215 pKa = 3.81LLFPNTGKK223 pKa = 10.49KK224 pKa = 10.06KK225 pKa = 10.43KK226 pKa = 10.23FSLFDD231 pKa = 3.52WEE233 pKa = 4.59CEE235 pKa = 3.78RR236 pKa = 11.84DD237 pKa = 3.69LACAFCRR244 pKa = 11.84PMRR247 pKa = 11.84FYY249 pKa = 10.71PSDD252 pKa = 3.43NPTYY256 pKa = 10.05PWLPPKK262 pKa = 10.24RR263 pKa = 11.84DD264 pKa = 3.07IPKK267 pKa = 8.76ICKK270 pKa = 9.96VNFKK274 pKa = 10.86INFTEE279 pKa = 3.93
MM1 pKa = 7.39PWSLPRR7 pKa = 11.84HH8 pKa = 5.86NIRR11 pKa = 11.84TRR13 pKa = 11.84EE14 pKa = 4.01DD15 pKa = 3.01LWVQSILYY23 pKa = 10.14SHH25 pKa = 7.23DD26 pKa = 3.69TFCGCDD32 pKa = 3.75NIPEE36 pKa = 4.6HH37 pKa = 6.14LTGLLGGVRR46 pKa = 11.84PAPPRR51 pKa = 11.84NPGPPTIRR59 pKa = 11.84SLPALPPAPEE69 pKa = 4.26PPEE72 pKa = 3.88EE73 pKa = 4.04PRR75 pKa = 11.84RR76 pKa = 11.84GGDD79 pKa = 3.05TDD81 pKa = 3.6GDD83 pKa = 3.75RR84 pKa = 11.84GEE86 pKa = 5.28DD87 pKa = 3.44GGDD90 pKa = 2.93AAGAYY95 pKa = 9.42EE96 pKa = 4.39PEE98 pKa = 4.29DD99 pKa = 4.0LEE101 pKa = 4.46EE102 pKa = 4.44LFAAAEE108 pKa = 4.02QDD110 pKa = 3.82DD111 pKa = 3.88MRR113 pKa = 11.84RR114 pKa = 11.84VQKK117 pKa = 9.78TPIRR121 pKa = 11.84HH122 pKa = 5.33SPPRR126 pKa = 11.84AAIKK130 pKa = 9.45PRR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84RR135 pKa = 11.84LVYY138 pKa = 9.86PQTTPKK144 pKa = 10.2RR145 pKa = 11.84ARR147 pKa = 11.84RR148 pKa = 11.84DD149 pKa = 3.42HH150 pKa = 6.42LRR152 pKa = 11.84GRR154 pKa = 11.84TSTPKK159 pKa = 10.15RR160 pKa = 11.84RR161 pKa = 11.84GAKK164 pKa = 9.46RR165 pKa = 11.84KK166 pKa = 10.12APTATQSPATAPASPQTGNQTPHH189 pKa = 5.98GRR191 pKa = 11.84RR192 pKa = 11.84PPTQTGSPLEE202 pKa = 4.17PSPIILPPEE211 pKa = 5.15PIPDD215 pKa = 3.81LLFPNTGKK223 pKa = 10.49KK224 pKa = 10.06KK225 pKa = 10.43KK226 pKa = 10.23FSLFDD231 pKa = 3.52WEE233 pKa = 4.59CEE235 pKa = 3.78RR236 pKa = 11.84DD237 pKa = 3.69LACAFCRR244 pKa = 11.84PMRR247 pKa = 11.84FYY249 pKa = 10.71PSDD252 pKa = 3.43NPTYY256 pKa = 10.05PWLPPKK262 pKa = 10.24RR263 pKa = 11.84DD264 pKa = 3.07IPKK267 pKa = 8.76ICKK270 pKa = 9.96VNFKK274 pKa = 10.86INFTEE279 pKa = 3.93
Molecular weight: 31.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1426 |
112 |
746 |
356.5 |
41.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.839 ± 0.878 | 1.753 ± 0.112 |
5.68 ± 1.029 | 5.54 ± 0.965 |
2.595 ± 0.439 | 5.68 ± 0.867 |
2.665 ± 0.491 | 4.628 ± 0.548 |
5.68 ± 0.856 | 7.223 ± 0.312 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.964 ± 0.478 | 4.278 ± 0.497 |
9.958 ± 2.487 | 4.208 ± 0.741 |
9.537 ± 0.725 | 6.522 ± 1.561 |
7.714 ± 0.732 | 2.735 ± 0.702 |
2.665 ± 0.733 | 4.137 ± 1.195 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
