
Planctomycetales bacterium 10988
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; unclassified Planctomycetales
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
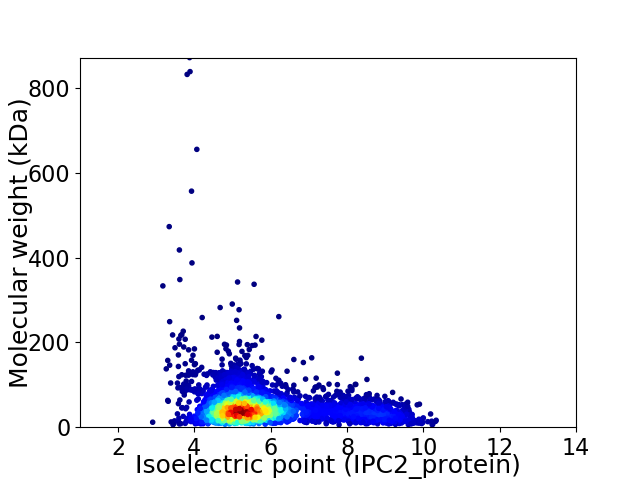
Virtual 2D-PAGE plot for 4056 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q3LFH9|A0A5Q3LFH9_9BACT Tripeptidyl-peptidase II OS=Planctomycetales bacterium 10988 OX=2603660 GN=PBC10988_40250 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 9.68WLAPFRR8 pKa = 11.84RR9 pKa = 11.84WQRR12 pKa = 11.84NKK14 pKa = 10.29SQQASAARR22 pKa = 11.84ITPSTPQRR30 pKa = 11.84PIRR33 pKa = 11.84RR34 pKa = 11.84CRR36 pKa = 11.84IEE38 pKa = 3.94AMEE41 pKa = 4.11RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 4.09MLSADD49 pKa = 3.84PLHH52 pKa = 6.72FGAVYY57 pKa = 10.87YY58 pKa = 10.68EE59 pKa = 4.08DD60 pKa = 5.29AFLSGADD67 pKa = 3.7FSNPGDD73 pKa = 4.02LIEE76 pKa = 4.24ITFEE80 pKa = 3.9GGAAGTQMHH89 pKa = 6.91SLTIDD94 pKa = 4.08LDD96 pKa = 4.13PNAGGLEE103 pKa = 4.08AGDD106 pKa = 4.61RR107 pKa = 11.84IFDD110 pKa = 3.92TLAGGLGSSGAVPLTIVEE128 pKa = 4.29NNGIDD133 pKa = 3.81SINFSLSSNDD143 pKa = 3.95LDD145 pKa = 4.0GQTSLTITFSGFDD158 pKa = 3.33VGDD161 pKa = 3.75SFTIGVDD168 pKa = 2.8IDD170 pKa = 3.82EE171 pKa = 4.43VAEE174 pKa = 4.6GGGLTPTVEE183 pKa = 4.11GAEE186 pKa = 4.38FEE188 pKa = 4.76GSTLTATFSADD199 pKa = 2.88TYY201 pKa = 11.26YY202 pKa = 11.32DD203 pKa = 3.44STKK206 pKa = 10.25TATFYY211 pKa = 11.45NQFSLDD217 pKa = 3.6GTGLQLPPDD226 pKa = 4.71DD227 pKa = 5.2YY228 pKa = 11.33IPPNSVPAPDD238 pKa = 3.72YY239 pKa = 8.79TAGAIGEE246 pKa = 4.3LQQVPLPITISGTVFEE262 pKa = 5.79DD263 pKa = 3.94GNLNNHH269 pKa = 5.94QDD271 pKa = 3.4GGEE274 pKa = 3.84QGIGGVEE281 pKa = 3.9LEE283 pKa = 3.91LWQWNGSQYY292 pKa = 11.01EE293 pKa = 4.25STGRR297 pKa = 11.84VTTTDD302 pKa = 2.96SQGDD306 pKa = 3.7YY307 pKa = 10.94HH308 pKa = 8.25FDD310 pKa = 3.55DD311 pKa = 4.5VDD313 pKa = 3.58PGRR316 pKa = 11.84YY317 pKa = 8.22RR318 pKa = 11.84VVEE321 pKa = 4.09HH322 pKa = 6.39QPDD325 pKa = 4.69GYY327 pKa = 10.56HH328 pKa = 6.41SVGAEE333 pKa = 3.57IGSVDD338 pKa = 3.53GSPRR342 pKa = 11.84GSVFSPDD349 pKa = 3.15ILTDD353 pKa = 3.37IQLVGGDD360 pKa = 3.85DD361 pKa = 3.75TIEE364 pKa = 4.12NNFAEE369 pKa = 4.51AAANSIEE376 pKa = 3.84GVVYY380 pKa = 10.44HH381 pKa = 7.36DD382 pKa = 4.52RR383 pKa = 11.84DD384 pKa = 3.32NDD386 pKa = 3.61GFQDD390 pKa = 3.48VGEE393 pKa = 4.21EE394 pKa = 4.54GIAGAEE400 pKa = 4.0VQLFDD405 pKa = 3.93QQGNLVATTFTNAQGEE421 pKa = 4.55YY422 pKa = 10.65LFDD425 pKa = 4.79DD426 pKa = 4.16LHH428 pKa = 6.67FGSYY432 pKa = 9.94SLQEE436 pKa = 4.04IQPAGYY442 pKa = 10.59LDD444 pKa = 4.16GLDD447 pKa = 3.49TAGSVGGLAEE457 pKa = 4.27NPGDD461 pKa = 5.11AIRR464 pKa = 11.84QIQLTSGTHH473 pKa = 4.38ATDD476 pKa = 3.46YY477 pKa = 11.29LFGEE481 pKa = 4.96LVPASISGQVFADD494 pKa = 4.17LDD496 pKa = 4.06GDD498 pKa = 4.37CYY500 pKa = 10.88FDD502 pKa = 4.37PEE504 pKa = 4.18EE505 pKa = 4.6SPIEE509 pKa = 3.93GVEE512 pKa = 3.41IHH514 pKa = 6.68LRR516 pKa = 11.84DD517 pKa = 3.57VQGNIVATTQTDD529 pKa = 3.41AQGLYY534 pKa = 10.22QFDD537 pKa = 3.67NLRR540 pKa = 11.84PGEE543 pKa = 4.21YY544 pKa = 8.52TVHH547 pKa = 6.06EE548 pKa = 4.36VQPFGYY554 pKa = 8.61YY555 pKa = 9.34QGGVVVGSAGGVISTFEE572 pKa = 3.93QDD574 pKa = 3.35TVTSIALVSGEE585 pKa = 3.94NAAGYY590 pKa = 6.59TFCEE594 pKa = 4.51VPPATLAGNVYY605 pKa = 10.45VDD607 pKa = 3.87ANQNGVRR614 pKa = 11.84DD615 pKa = 3.99GGEE618 pKa = 3.67QGIGGVTLEE627 pKa = 4.57LRR629 pKa = 11.84DD630 pKa = 3.69AGGSVVATTATDD642 pKa = 3.29STGAYY647 pKa = 9.89LFDD650 pKa = 5.36SILPGEE656 pKa = 4.33YY657 pKa = 9.34SVHH660 pKa = 5.55EE661 pKa = 4.26VQPFGYY667 pKa = 10.05FDD669 pKa = 3.96GEE671 pKa = 4.21EE672 pKa = 4.42SIGTAGGVLGPGNDD686 pKa = 3.65VISQVTLHH694 pKa = 6.54PAEE697 pKa = 4.89HH698 pKa = 5.35ATEE701 pKa = 4.28YY702 pKa = 11.29NFGEE706 pKa = 4.63LLPASIQGSVFADD719 pKa = 3.63LDD721 pKa = 3.97GDD723 pKa = 4.79CIQDD727 pKa = 3.69PGEE730 pKa = 4.39SPLEE734 pKa = 3.82NVVIQLLNGNGDD746 pKa = 3.99VIDD749 pKa = 4.06STVTDD754 pKa = 3.82ANGDD758 pKa = 3.64YY759 pKa = 10.68SFEE762 pKa = 4.16SLPPGTYY769 pKa = 10.26GIRR772 pKa = 11.84EE773 pKa = 4.14IQPTGYY779 pKa = 9.62FQGGTVVGSAGGTLAPGDD797 pKa = 4.22ADD799 pKa = 3.77TIVAIEE805 pKa = 4.0LHH807 pKa = 6.27SGEE810 pKa = 4.55AATEE814 pKa = 4.17YY815 pKa = 10.53VFCEE819 pKa = 4.22HH820 pKa = 7.4PPAQLSGEE828 pKa = 4.4VYY830 pKa = 10.67VDD832 pKa = 3.5QNQSGTLDD840 pKa = 3.3SGEE843 pKa = 4.08RR844 pKa = 11.84GIGGVTLEE852 pKa = 4.45LLDD855 pKa = 4.06VNGQVVATTVTNDD868 pKa = 2.92AGHH871 pKa = 6.3YY872 pKa = 10.55LFDD875 pKa = 3.68QLPPGTYY882 pKa = 9.83SIRR885 pKa = 11.84EE886 pKa = 4.05IQPAGYY892 pKa = 10.12FDD894 pKa = 6.11GIDD897 pKa = 3.33QAGSLGGIAQNPGDD911 pKa = 4.23QISQIVLGPGMQGVEE926 pKa = 4.06YY927 pKa = 10.56RR928 pKa = 11.84FGEE931 pKa = 4.19YY932 pKa = 10.31EE933 pKa = 3.88PASLHH938 pKa = 5.65GRR940 pKa = 11.84VFADD944 pKa = 4.57LDD946 pKa = 3.98GDD948 pKa = 4.71CILDD952 pKa = 3.85ANEE955 pKa = 3.88TPIAGVQIEE964 pKa = 4.57LLDD967 pKa = 3.65GTGQVIATTTTDD979 pKa = 2.71AGGDD983 pKa = 3.62YY984 pKa = 10.42SFEE987 pKa = 4.06GLPPGTYY994 pKa = 9.5GVRR997 pKa = 11.84EE998 pKa = 4.06IQPAGYY1004 pKa = 8.56YY1005 pKa = 9.86QGGTMVGTAGGTLSSGDD1022 pKa = 3.48SDD1024 pKa = 4.1TIVAIEE1030 pKa = 4.4LGSGTVATEE1039 pKa = 4.12YY1040 pKa = 10.68VFCEE1044 pKa = 4.12HH1045 pKa = 7.18PPSKK1049 pKa = 10.45LAGSVYY1055 pKa = 10.71VDD1057 pKa = 3.81ANQNGNFDD1065 pKa = 3.65SGEE1068 pKa = 3.96TGIAGVEE1075 pKa = 3.88LEE1077 pKa = 5.05LYY1079 pKa = 10.17NSNGDD1084 pKa = 3.63LVATVVTDD1092 pKa = 3.32EE1093 pKa = 4.12TGFYY1097 pKa = 10.45CFEE1100 pKa = 4.35NLPPDD1105 pKa = 3.87TYY1107 pKa = 10.64TIEE1110 pKa = 4.39EE1111 pKa = 4.27VQPPGYY1117 pKa = 10.0YY1118 pKa = 10.35DD1119 pKa = 4.46GVDD1122 pKa = 3.3TPGTLGGIPEE1132 pKa = 4.39NPGDD1136 pKa = 4.4SIRR1139 pKa = 11.84GIALPPGTTGEE1150 pKa = 4.11EE1151 pKa = 4.7YY1152 pKa = 11.34VFGEE1156 pKa = 4.33VLAGRR1161 pKa = 11.84ISGRR1165 pKa = 11.84VHH1167 pKa = 7.08ADD1169 pKa = 3.04LDD1171 pKa = 4.13GDD1173 pKa = 4.64CIFDD1177 pKa = 4.03PEE1179 pKa = 4.21EE1180 pKa = 4.48SPLVGVEE1187 pKa = 4.79IEE1189 pKa = 4.51LLDD1192 pKa = 4.63DD1193 pKa = 4.86AGQVVATTFTNDD1205 pKa = 2.77AGEE1208 pKa = 4.16YY1209 pKa = 10.57LFDD1212 pKa = 4.13NLPPGTYY1219 pKa = 8.97SVRR1222 pKa = 11.84EE1223 pKa = 3.84IQPAGFFDD1231 pKa = 4.06GMASVGSGTGTVTDD1245 pKa = 4.38SNTIAAISIGSGQTLVDD1262 pKa = 3.47YY1263 pKa = 9.62DD1264 pKa = 4.0FCEE1267 pKa = 4.4EE1268 pKa = 3.97PHH1270 pKa = 6.62ASLSGRR1276 pKa = 11.84VYY1278 pKa = 10.59QDD1280 pKa = 2.88GPNIVLSAGTTIDD1293 pKa = 4.43LSNTRR1298 pKa = 11.84DD1299 pKa = 4.02GILSADD1305 pKa = 3.85DD1306 pKa = 3.58TPIAGVTLILGDD1318 pKa = 3.83TNGNPILDD1326 pKa = 3.52ASGNLITTVTGSDD1339 pKa = 2.99GSFRR1343 pKa = 11.84FDD1345 pKa = 3.15ALHH1348 pKa = 6.45AGSYY1352 pKa = 10.82SLLQVQPDD1360 pKa = 3.96DD1361 pKa = 3.92YY1362 pKa = 11.72VDD1364 pKa = 3.81NRR1366 pKa = 11.84DD1367 pKa = 3.42HH1368 pKa = 7.34AGDD1371 pKa = 3.75AGGTAVNPGDD1381 pKa = 4.7AILGIEE1387 pKa = 4.79LKK1389 pKa = 9.88TGQQASLYY1397 pKa = 9.32HH1398 pKa = 5.95FSEE1401 pKa = 4.97LKK1403 pKa = 10.78VEE1405 pKa = 4.37TPPGLIIRR1413 pKa = 11.84PPLDD1417 pKa = 3.99PLPSLGGPVLVGAPPAPAPIIVAPPVVSVNRR1448 pKa = 11.84VDD1450 pKa = 4.58LGAGLGVGNAGDD1462 pKa = 4.39GYY1464 pKa = 8.06TWHH1467 pKa = 7.22LSVIDD1472 pKa = 3.72GGFPRR1477 pKa = 11.84GNVSTASADD1486 pKa = 3.53NVIRR1490 pKa = 11.84PVYY1493 pKa = 10.03FSPEE1497 pKa = 3.7TWEE1500 pKa = 4.69GPDD1503 pKa = 3.21MTEE1506 pKa = 5.36SLWQLPSGTDD1516 pKa = 3.18ALIVDD1521 pKa = 4.46RR1522 pKa = 11.84SFAFGPLEE1530 pKa = 4.48GIPVTGDD1537 pKa = 3.19FNGDD1541 pKa = 3.09GVTDD1545 pKa = 3.31IGIFVDD1551 pKa = 4.74GEE1553 pKa = 4.03WFIDD1557 pKa = 3.52ANGNGRR1563 pKa = 11.84WDD1565 pKa = 4.35EE1566 pKa = 3.87GDD1568 pKa = 2.95MWAKK1572 pKa = 10.21LGKK1575 pKa = 9.61PGDD1578 pKa = 3.88RR1579 pKa = 11.84PVVGDD1584 pKa = 3.48WNGDD1588 pKa = 3.35GKK1590 pKa = 11.35SDD1592 pKa = 3.08IGVYY1596 pKa = 9.96GLPWEE1601 pKa = 4.73GDD1603 pKa = 3.44DD1604 pKa = 3.76RR1605 pKa = 11.84ALAAEE1610 pKa = 4.27VGLPSAEE1617 pKa = 3.97NPPIGVQKK1625 pKa = 10.52NVPPRR1630 pKa = 11.84QDD1632 pKa = 3.34EE1633 pKa = 4.48APVTPRR1639 pKa = 11.84LLKK1642 pKa = 9.13HH1643 pKa = 5.18TAAGEE1648 pKa = 4.14VRR1650 pKa = 11.84ADD1652 pKa = 4.39LIDD1655 pKa = 3.61HH1656 pKa = 5.92VFAYY1660 pKa = 9.6GQAGDD1665 pKa = 4.34IPLSGDD1671 pKa = 3.03WNGDD1675 pKa = 3.02GMDD1678 pKa = 3.2KK1679 pKa = 10.99VGVYY1683 pKa = 10.55RR1684 pKa = 11.84NGIWYY1689 pKa = 9.93LDD1691 pKa = 3.55VDD1693 pKa = 3.94GDD1695 pKa = 4.36GQFTATDD1702 pKa = 3.73EE1703 pKa = 4.35VVHH1706 pKa = 7.34LGEE1709 pKa = 5.35DD1710 pKa = 3.09VGLPIVGDD1718 pKa = 3.79FDD1720 pKa = 5.51GDD1722 pKa = 4.08GVDD1725 pKa = 4.64EE1726 pKa = 4.94LGLYY1730 pKa = 9.44QEE1732 pKa = 5.2GVWKK1736 pKa = 10.29IDD1738 pKa = 3.41TNGDD1742 pKa = 3.59HH1743 pKa = 7.07QLDD1746 pKa = 3.85ALDD1749 pKa = 4.79KK1750 pKa = 9.68VFEE1753 pKa = 4.66LGGPEE1758 pKa = 4.05SLPIVGDD1765 pKa = 3.57FDD1767 pKa = 5.24GDD1769 pKa = 4.0GIEE1772 pKa = 4.21EE1773 pKa = 4.15VGLYY1777 pKa = 10.23RR1778 pKa = 11.84PGAGYY1783 pKa = 9.96DD1784 pKa = 3.69ANSEE1788 pKa = 4.0AAQDD1792 pKa = 3.29HH1793 pKa = 6.91DD1794 pKa = 3.94VAQRR1798 pKa = 11.84EE1799 pKa = 4.43SSRR1802 pKa = 11.84KK1803 pKa = 9.66RR1804 pKa = 11.84II1805 pKa = 3.64
MM1 pKa = 7.37KK2 pKa = 9.68WLAPFRR8 pKa = 11.84RR9 pKa = 11.84WQRR12 pKa = 11.84NKK14 pKa = 10.29SQQASAARR22 pKa = 11.84ITPSTPQRR30 pKa = 11.84PIRR33 pKa = 11.84RR34 pKa = 11.84CRR36 pKa = 11.84IEE38 pKa = 3.94AMEE41 pKa = 4.11RR42 pKa = 11.84RR43 pKa = 11.84EE44 pKa = 4.09MLSADD49 pKa = 3.84PLHH52 pKa = 6.72FGAVYY57 pKa = 10.87YY58 pKa = 10.68EE59 pKa = 4.08DD60 pKa = 5.29AFLSGADD67 pKa = 3.7FSNPGDD73 pKa = 4.02LIEE76 pKa = 4.24ITFEE80 pKa = 3.9GGAAGTQMHH89 pKa = 6.91SLTIDD94 pKa = 4.08LDD96 pKa = 4.13PNAGGLEE103 pKa = 4.08AGDD106 pKa = 4.61RR107 pKa = 11.84IFDD110 pKa = 3.92TLAGGLGSSGAVPLTIVEE128 pKa = 4.29NNGIDD133 pKa = 3.81SINFSLSSNDD143 pKa = 3.95LDD145 pKa = 4.0GQTSLTITFSGFDD158 pKa = 3.33VGDD161 pKa = 3.75SFTIGVDD168 pKa = 2.8IDD170 pKa = 3.82EE171 pKa = 4.43VAEE174 pKa = 4.6GGGLTPTVEE183 pKa = 4.11GAEE186 pKa = 4.38FEE188 pKa = 4.76GSTLTATFSADD199 pKa = 2.88TYY201 pKa = 11.26YY202 pKa = 11.32DD203 pKa = 3.44STKK206 pKa = 10.25TATFYY211 pKa = 11.45NQFSLDD217 pKa = 3.6GTGLQLPPDD226 pKa = 4.71DD227 pKa = 5.2YY228 pKa = 11.33IPPNSVPAPDD238 pKa = 3.72YY239 pKa = 8.79TAGAIGEE246 pKa = 4.3LQQVPLPITISGTVFEE262 pKa = 5.79DD263 pKa = 3.94GNLNNHH269 pKa = 5.94QDD271 pKa = 3.4GGEE274 pKa = 3.84QGIGGVEE281 pKa = 3.9LEE283 pKa = 3.91LWQWNGSQYY292 pKa = 11.01EE293 pKa = 4.25STGRR297 pKa = 11.84VTTTDD302 pKa = 2.96SQGDD306 pKa = 3.7YY307 pKa = 10.94HH308 pKa = 8.25FDD310 pKa = 3.55DD311 pKa = 4.5VDD313 pKa = 3.58PGRR316 pKa = 11.84YY317 pKa = 8.22RR318 pKa = 11.84VVEE321 pKa = 4.09HH322 pKa = 6.39QPDD325 pKa = 4.69GYY327 pKa = 10.56HH328 pKa = 6.41SVGAEE333 pKa = 3.57IGSVDD338 pKa = 3.53GSPRR342 pKa = 11.84GSVFSPDD349 pKa = 3.15ILTDD353 pKa = 3.37IQLVGGDD360 pKa = 3.85DD361 pKa = 3.75TIEE364 pKa = 4.12NNFAEE369 pKa = 4.51AAANSIEE376 pKa = 3.84GVVYY380 pKa = 10.44HH381 pKa = 7.36DD382 pKa = 4.52RR383 pKa = 11.84DD384 pKa = 3.32NDD386 pKa = 3.61GFQDD390 pKa = 3.48VGEE393 pKa = 4.21EE394 pKa = 4.54GIAGAEE400 pKa = 4.0VQLFDD405 pKa = 3.93QQGNLVATTFTNAQGEE421 pKa = 4.55YY422 pKa = 10.65LFDD425 pKa = 4.79DD426 pKa = 4.16LHH428 pKa = 6.67FGSYY432 pKa = 9.94SLQEE436 pKa = 4.04IQPAGYY442 pKa = 10.59LDD444 pKa = 4.16GLDD447 pKa = 3.49TAGSVGGLAEE457 pKa = 4.27NPGDD461 pKa = 5.11AIRR464 pKa = 11.84QIQLTSGTHH473 pKa = 4.38ATDD476 pKa = 3.46YY477 pKa = 11.29LFGEE481 pKa = 4.96LVPASISGQVFADD494 pKa = 4.17LDD496 pKa = 4.06GDD498 pKa = 4.37CYY500 pKa = 10.88FDD502 pKa = 4.37PEE504 pKa = 4.18EE505 pKa = 4.6SPIEE509 pKa = 3.93GVEE512 pKa = 3.41IHH514 pKa = 6.68LRR516 pKa = 11.84DD517 pKa = 3.57VQGNIVATTQTDD529 pKa = 3.41AQGLYY534 pKa = 10.22QFDD537 pKa = 3.67NLRR540 pKa = 11.84PGEE543 pKa = 4.21YY544 pKa = 8.52TVHH547 pKa = 6.06EE548 pKa = 4.36VQPFGYY554 pKa = 8.61YY555 pKa = 9.34QGGVVVGSAGGVISTFEE572 pKa = 3.93QDD574 pKa = 3.35TVTSIALVSGEE585 pKa = 3.94NAAGYY590 pKa = 6.59TFCEE594 pKa = 4.51VPPATLAGNVYY605 pKa = 10.45VDD607 pKa = 3.87ANQNGVRR614 pKa = 11.84DD615 pKa = 3.99GGEE618 pKa = 3.67QGIGGVTLEE627 pKa = 4.57LRR629 pKa = 11.84DD630 pKa = 3.69AGGSVVATTATDD642 pKa = 3.29STGAYY647 pKa = 9.89LFDD650 pKa = 5.36SILPGEE656 pKa = 4.33YY657 pKa = 9.34SVHH660 pKa = 5.55EE661 pKa = 4.26VQPFGYY667 pKa = 10.05FDD669 pKa = 3.96GEE671 pKa = 4.21EE672 pKa = 4.42SIGTAGGVLGPGNDD686 pKa = 3.65VISQVTLHH694 pKa = 6.54PAEE697 pKa = 4.89HH698 pKa = 5.35ATEE701 pKa = 4.28YY702 pKa = 11.29NFGEE706 pKa = 4.63LLPASIQGSVFADD719 pKa = 3.63LDD721 pKa = 3.97GDD723 pKa = 4.79CIQDD727 pKa = 3.69PGEE730 pKa = 4.39SPLEE734 pKa = 3.82NVVIQLLNGNGDD746 pKa = 3.99VIDD749 pKa = 4.06STVTDD754 pKa = 3.82ANGDD758 pKa = 3.64YY759 pKa = 10.68SFEE762 pKa = 4.16SLPPGTYY769 pKa = 10.26GIRR772 pKa = 11.84EE773 pKa = 4.14IQPTGYY779 pKa = 9.62FQGGTVVGSAGGTLAPGDD797 pKa = 4.22ADD799 pKa = 3.77TIVAIEE805 pKa = 4.0LHH807 pKa = 6.27SGEE810 pKa = 4.55AATEE814 pKa = 4.17YY815 pKa = 10.53VFCEE819 pKa = 4.22HH820 pKa = 7.4PPAQLSGEE828 pKa = 4.4VYY830 pKa = 10.67VDD832 pKa = 3.5QNQSGTLDD840 pKa = 3.3SGEE843 pKa = 4.08RR844 pKa = 11.84GIGGVTLEE852 pKa = 4.45LLDD855 pKa = 4.06VNGQVVATTVTNDD868 pKa = 2.92AGHH871 pKa = 6.3YY872 pKa = 10.55LFDD875 pKa = 3.68QLPPGTYY882 pKa = 9.83SIRR885 pKa = 11.84EE886 pKa = 4.05IQPAGYY892 pKa = 10.12FDD894 pKa = 6.11GIDD897 pKa = 3.33QAGSLGGIAQNPGDD911 pKa = 4.23QISQIVLGPGMQGVEE926 pKa = 4.06YY927 pKa = 10.56RR928 pKa = 11.84FGEE931 pKa = 4.19YY932 pKa = 10.31EE933 pKa = 3.88PASLHH938 pKa = 5.65GRR940 pKa = 11.84VFADD944 pKa = 4.57LDD946 pKa = 3.98GDD948 pKa = 4.71CILDD952 pKa = 3.85ANEE955 pKa = 3.88TPIAGVQIEE964 pKa = 4.57LLDD967 pKa = 3.65GTGQVIATTTTDD979 pKa = 2.71AGGDD983 pKa = 3.62YY984 pKa = 10.42SFEE987 pKa = 4.06GLPPGTYY994 pKa = 9.5GVRR997 pKa = 11.84EE998 pKa = 4.06IQPAGYY1004 pKa = 8.56YY1005 pKa = 9.86QGGTMVGTAGGTLSSGDD1022 pKa = 3.48SDD1024 pKa = 4.1TIVAIEE1030 pKa = 4.4LGSGTVATEE1039 pKa = 4.12YY1040 pKa = 10.68VFCEE1044 pKa = 4.12HH1045 pKa = 7.18PPSKK1049 pKa = 10.45LAGSVYY1055 pKa = 10.71VDD1057 pKa = 3.81ANQNGNFDD1065 pKa = 3.65SGEE1068 pKa = 3.96TGIAGVEE1075 pKa = 3.88LEE1077 pKa = 5.05LYY1079 pKa = 10.17NSNGDD1084 pKa = 3.63LVATVVTDD1092 pKa = 3.32EE1093 pKa = 4.12TGFYY1097 pKa = 10.45CFEE1100 pKa = 4.35NLPPDD1105 pKa = 3.87TYY1107 pKa = 10.64TIEE1110 pKa = 4.39EE1111 pKa = 4.27VQPPGYY1117 pKa = 10.0YY1118 pKa = 10.35DD1119 pKa = 4.46GVDD1122 pKa = 3.3TPGTLGGIPEE1132 pKa = 4.39NPGDD1136 pKa = 4.4SIRR1139 pKa = 11.84GIALPPGTTGEE1150 pKa = 4.11EE1151 pKa = 4.7YY1152 pKa = 11.34VFGEE1156 pKa = 4.33VLAGRR1161 pKa = 11.84ISGRR1165 pKa = 11.84VHH1167 pKa = 7.08ADD1169 pKa = 3.04LDD1171 pKa = 4.13GDD1173 pKa = 4.64CIFDD1177 pKa = 4.03PEE1179 pKa = 4.21EE1180 pKa = 4.48SPLVGVEE1187 pKa = 4.79IEE1189 pKa = 4.51LLDD1192 pKa = 4.63DD1193 pKa = 4.86AGQVVATTFTNDD1205 pKa = 2.77AGEE1208 pKa = 4.16YY1209 pKa = 10.57LFDD1212 pKa = 4.13NLPPGTYY1219 pKa = 8.97SVRR1222 pKa = 11.84EE1223 pKa = 3.84IQPAGFFDD1231 pKa = 4.06GMASVGSGTGTVTDD1245 pKa = 4.38SNTIAAISIGSGQTLVDD1262 pKa = 3.47YY1263 pKa = 9.62DD1264 pKa = 4.0FCEE1267 pKa = 4.4EE1268 pKa = 3.97PHH1270 pKa = 6.62ASLSGRR1276 pKa = 11.84VYY1278 pKa = 10.59QDD1280 pKa = 2.88GPNIVLSAGTTIDD1293 pKa = 4.43LSNTRR1298 pKa = 11.84DD1299 pKa = 4.02GILSADD1305 pKa = 3.85DD1306 pKa = 3.58TPIAGVTLILGDD1318 pKa = 3.83TNGNPILDD1326 pKa = 3.52ASGNLITTVTGSDD1339 pKa = 2.99GSFRR1343 pKa = 11.84FDD1345 pKa = 3.15ALHH1348 pKa = 6.45AGSYY1352 pKa = 10.82SLLQVQPDD1360 pKa = 3.96DD1361 pKa = 3.92YY1362 pKa = 11.72VDD1364 pKa = 3.81NRR1366 pKa = 11.84DD1367 pKa = 3.42HH1368 pKa = 7.34AGDD1371 pKa = 3.75AGGTAVNPGDD1381 pKa = 4.7AILGIEE1387 pKa = 4.79LKK1389 pKa = 9.88TGQQASLYY1397 pKa = 9.32HH1398 pKa = 5.95FSEE1401 pKa = 4.97LKK1403 pKa = 10.78VEE1405 pKa = 4.37TPPGLIIRR1413 pKa = 11.84PPLDD1417 pKa = 3.99PLPSLGGPVLVGAPPAPAPIIVAPPVVSVNRR1448 pKa = 11.84VDD1450 pKa = 4.58LGAGLGVGNAGDD1462 pKa = 4.39GYY1464 pKa = 8.06TWHH1467 pKa = 7.22LSVIDD1472 pKa = 3.72GGFPRR1477 pKa = 11.84GNVSTASADD1486 pKa = 3.53NVIRR1490 pKa = 11.84PVYY1493 pKa = 10.03FSPEE1497 pKa = 3.7TWEE1500 pKa = 4.69GPDD1503 pKa = 3.21MTEE1506 pKa = 5.36SLWQLPSGTDD1516 pKa = 3.18ALIVDD1521 pKa = 4.46RR1522 pKa = 11.84SFAFGPLEE1530 pKa = 4.48GIPVTGDD1537 pKa = 3.19FNGDD1541 pKa = 3.09GVTDD1545 pKa = 3.31IGIFVDD1551 pKa = 4.74GEE1553 pKa = 4.03WFIDD1557 pKa = 3.52ANGNGRR1563 pKa = 11.84WDD1565 pKa = 4.35EE1566 pKa = 3.87GDD1568 pKa = 2.95MWAKK1572 pKa = 10.21LGKK1575 pKa = 9.61PGDD1578 pKa = 3.88RR1579 pKa = 11.84PVVGDD1584 pKa = 3.48WNGDD1588 pKa = 3.35GKK1590 pKa = 11.35SDD1592 pKa = 3.08IGVYY1596 pKa = 9.96GLPWEE1601 pKa = 4.73GDD1603 pKa = 3.44DD1604 pKa = 3.76RR1605 pKa = 11.84ALAAEE1610 pKa = 4.27VGLPSAEE1617 pKa = 3.97NPPIGVQKK1625 pKa = 10.52NVPPRR1630 pKa = 11.84QDD1632 pKa = 3.34EE1633 pKa = 4.48APVTPRR1639 pKa = 11.84LLKK1642 pKa = 9.13HH1643 pKa = 5.18TAAGEE1648 pKa = 4.14VRR1650 pKa = 11.84ADD1652 pKa = 4.39LIDD1655 pKa = 3.61HH1656 pKa = 5.92VFAYY1660 pKa = 9.6GQAGDD1665 pKa = 4.34IPLSGDD1671 pKa = 3.03WNGDD1675 pKa = 3.02GMDD1678 pKa = 3.2KK1679 pKa = 10.99VGVYY1683 pKa = 10.55RR1684 pKa = 11.84NGIWYY1689 pKa = 9.93LDD1691 pKa = 3.55VDD1693 pKa = 3.94GDD1695 pKa = 4.36GQFTATDD1702 pKa = 3.73EE1703 pKa = 4.35VVHH1706 pKa = 7.34LGEE1709 pKa = 5.35DD1710 pKa = 3.09VGLPIVGDD1718 pKa = 3.79FDD1720 pKa = 5.51GDD1722 pKa = 4.08GVDD1725 pKa = 4.64EE1726 pKa = 4.94LGLYY1730 pKa = 9.44QEE1732 pKa = 5.2GVWKK1736 pKa = 10.29IDD1738 pKa = 3.41TNGDD1742 pKa = 3.59HH1743 pKa = 7.07QLDD1746 pKa = 3.85ALDD1749 pKa = 4.79KK1750 pKa = 9.68VFEE1753 pKa = 4.66LGGPEE1758 pKa = 4.05SLPIVGDD1765 pKa = 3.57FDD1767 pKa = 5.24GDD1769 pKa = 4.0GIEE1772 pKa = 4.21EE1773 pKa = 4.15VGLYY1777 pKa = 10.23RR1778 pKa = 11.84PGAGYY1783 pKa = 9.96DD1784 pKa = 3.69ANSEE1788 pKa = 4.0AAQDD1792 pKa = 3.29HH1793 pKa = 6.91DD1794 pKa = 3.94VAQRR1798 pKa = 11.84EE1799 pKa = 4.43SSRR1802 pKa = 11.84KK1803 pKa = 9.66RR1804 pKa = 11.84II1805 pKa = 3.64
Molecular weight: 189.37 kDa
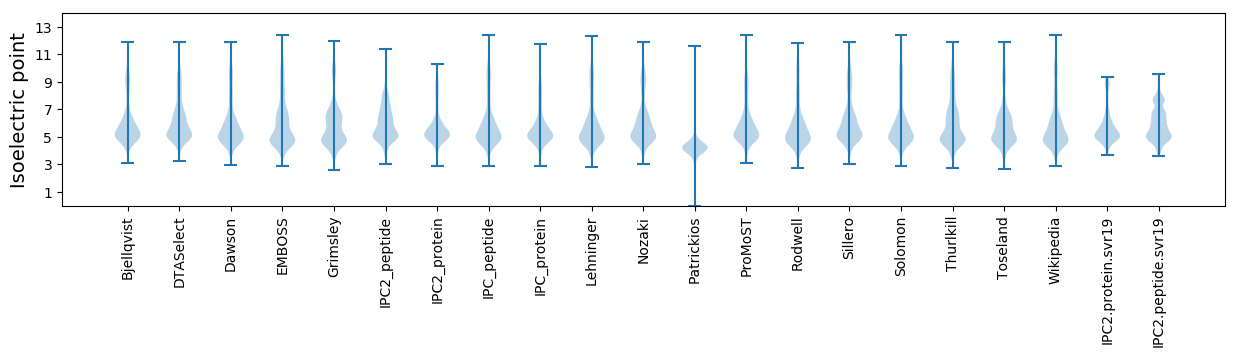
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q3L391|A0A5Q3L391_9BACT Dehydrogenase OS=Planctomycetales bacterium 10988 OX=2603660 GN=PBC10988_5230 PE=3 SV=1
MM1 pKa = 6.91NTIIPLSDD9 pKa = 3.45EE10 pKa = 4.01QRR12 pKa = 11.84KK13 pKa = 8.48QLLNVYY19 pKa = 10.25RR20 pKa = 11.84SGTDD24 pKa = 2.96PRR26 pKa = 11.84MSRR29 pKa = 11.84RR30 pKa = 11.84AHH32 pKa = 5.47VLLLFADD39 pKa = 4.29GWTWQQIRR47 pKa = 11.84KK48 pKa = 9.29ALFCSNDD55 pKa = 3.45LIDD58 pKa = 3.32RR59 pKa = 11.84TRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84FHH65 pKa = 6.68HH66 pKa = 6.34GGVPAVLQRR75 pKa = 11.84SDD77 pKa = 3.99ALPAASTSPGWWQRR91 pKa = 11.84VLKK94 pKa = 9.58WLTEE98 pKa = 4.07TTPGDD103 pKa = 3.28FGYY106 pKa = 10.46FRR108 pKa = 11.84SCWSCGLLAGLLAWEE123 pKa = 4.53TGVHH127 pKa = 6.51RR128 pKa = 11.84SAEE131 pKa = 4.17TIRR134 pKa = 11.84RR135 pKa = 11.84GLHH138 pKa = 5.88RR139 pKa = 11.84EE140 pKa = 3.46GDD142 pKa = 3.35VWRR145 pKa = 11.84RR146 pKa = 11.84PRR148 pKa = 11.84PVVGPTDD155 pKa = 3.58PDD157 pKa = 3.57YY158 pKa = 11.11SQKK161 pKa = 10.42LRR163 pKa = 11.84EE164 pKa = 4.16IRR166 pKa = 11.84HH167 pKa = 6.4LLDD170 pKa = 3.13QLPPSEE176 pKa = 4.11VAVFQDD182 pKa = 3.58EE183 pKa = 4.37VDD185 pKa = 3.66VNLNPKK191 pKa = 9.17IGPMWTPRR199 pKa = 11.84GQQAAVATPGNNEE212 pKa = 3.21KK213 pKa = 10.53RR214 pKa = 11.84YY215 pKa = 9.63LAGSLSWQTGRR226 pKa = 11.84LFVSPPQRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84NADD239 pKa = 3.3LFLSHH244 pKa = 7.29LAEE247 pKa = 4.19LQQRR251 pKa = 11.84LRR253 pKa = 11.84RR254 pKa = 11.84YY255 pKa = 9.17RR256 pKa = 11.84VIHH259 pKa = 5.31VLCDD263 pKa = 3.04NAKK266 pKa = 9.48FHH268 pKa = 6.72NCRR271 pKa = 11.84KK272 pKa = 9.36VRR274 pKa = 11.84QALQQRR280 pKa = 11.84LGRR283 pKa = 11.84IRR285 pKa = 11.84LHH287 pKa = 6.45FLPKK291 pKa = 9.97YY292 pKa = 10.45APEE295 pKa = 4.12TNPIEE300 pKa = 4.27RR301 pKa = 11.84VWWHH305 pKa = 4.92LHH307 pKa = 3.84EE308 pKa = 5.17TITRR312 pKa = 11.84NHH314 pKa = 6.18RR315 pKa = 11.84CEE317 pKa = 4.0TLEE320 pKa = 3.72EE321 pKa = 4.44LLRR324 pKa = 11.84QTCSGG329 pKa = 3.43
MM1 pKa = 6.91NTIIPLSDD9 pKa = 3.45EE10 pKa = 4.01QRR12 pKa = 11.84KK13 pKa = 8.48QLLNVYY19 pKa = 10.25RR20 pKa = 11.84SGTDD24 pKa = 2.96PRR26 pKa = 11.84MSRR29 pKa = 11.84RR30 pKa = 11.84AHH32 pKa = 5.47VLLLFADD39 pKa = 4.29GWTWQQIRR47 pKa = 11.84KK48 pKa = 9.29ALFCSNDD55 pKa = 3.45LIDD58 pKa = 3.32RR59 pKa = 11.84TRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84FHH65 pKa = 6.68HH66 pKa = 6.34GGVPAVLQRR75 pKa = 11.84SDD77 pKa = 3.99ALPAASTSPGWWQRR91 pKa = 11.84VLKK94 pKa = 9.58WLTEE98 pKa = 4.07TTPGDD103 pKa = 3.28FGYY106 pKa = 10.46FRR108 pKa = 11.84SCWSCGLLAGLLAWEE123 pKa = 4.53TGVHH127 pKa = 6.51RR128 pKa = 11.84SAEE131 pKa = 4.17TIRR134 pKa = 11.84RR135 pKa = 11.84GLHH138 pKa = 5.88RR139 pKa = 11.84EE140 pKa = 3.46GDD142 pKa = 3.35VWRR145 pKa = 11.84RR146 pKa = 11.84PRR148 pKa = 11.84PVVGPTDD155 pKa = 3.58PDD157 pKa = 3.57YY158 pKa = 11.11SQKK161 pKa = 10.42LRR163 pKa = 11.84EE164 pKa = 4.16IRR166 pKa = 11.84HH167 pKa = 6.4LLDD170 pKa = 3.13QLPPSEE176 pKa = 4.11VAVFQDD182 pKa = 3.58EE183 pKa = 4.37VDD185 pKa = 3.66VNLNPKK191 pKa = 9.17IGPMWTPRR199 pKa = 11.84GQQAAVATPGNNEE212 pKa = 3.21KK213 pKa = 10.53RR214 pKa = 11.84YY215 pKa = 9.63LAGSLSWQTGRR226 pKa = 11.84LFVSPPQRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84NADD239 pKa = 3.3LFLSHH244 pKa = 7.29LAEE247 pKa = 4.19LQQRR251 pKa = 11.84LRR253 pKa = 11.84RR254 pKa = 11.84YY255 pKa = 9.17RR256 pKa = 11.84VIHH259 pKa = 5.31VLCDD263 pKa = 3.04NAKK266 pKa = 9.48FHH268 pKa = 6.72NCRR271 pKa = 11.84KK272 pKa = 9.36VRR274 pKa = 11.84QALQQRR280 pKa = 11.84LGRR283 pKa = 11.84IRR285 pKa = 11.84LHH287 pKa = 6.45FLPKK291 pKa = 9.97YY292 pKa = 10.45APEE295 pKa = 4.12TNPIEE300 pKa = 4.27RR301 pKa = 11.84VWWHH305 pKa = 4.92LHH307 pKa = 3.84EE308 pKa = 5.17TITRR312 pKa = 11.84NHH314 pKa = 6.18RR315 pKa = 11.84CEE317 pKa = 4.0TLEE320 pKa = 3.72EE321 pKa = 4.44LLRR324 pKa = 11.84QTCSGG329 pKa = 3.43
Molecular weight: 38.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1770095 |
50 |
8529 |
436.4 |
48.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.21 ± 0.039 | 0.993 ± 0.015 |
5.582 ± 0.043 | 7.651 ± 0.047 |
4.1 ± 0.025 | 7.288 ± 0.051 |
2.152 ± 0.018 | 5.175 ± 0.026 |
4.306 ± 0.042 | 10.393 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.089 ± 0.017 | 3.373 ± 0.033 |
5.433 ± 0.033 | 4.707 ± 0.028 |
5.803 ± 0.045 | 6.493 ± 0.033 |
5.555 ± 0.044 | 6.296 ± 0.029 |
1.59 ± 0.018 | 2.813 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
