
Corynebacterium provencense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Corynebacteriaceae; Corynebacterium
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
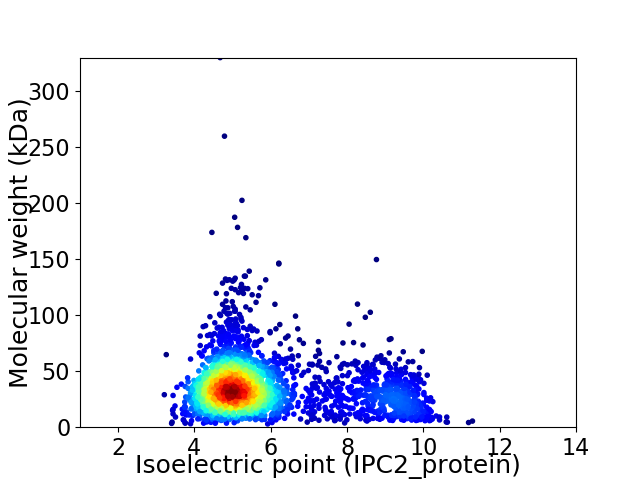
Virtual 2D-PAGE plot for 2661 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
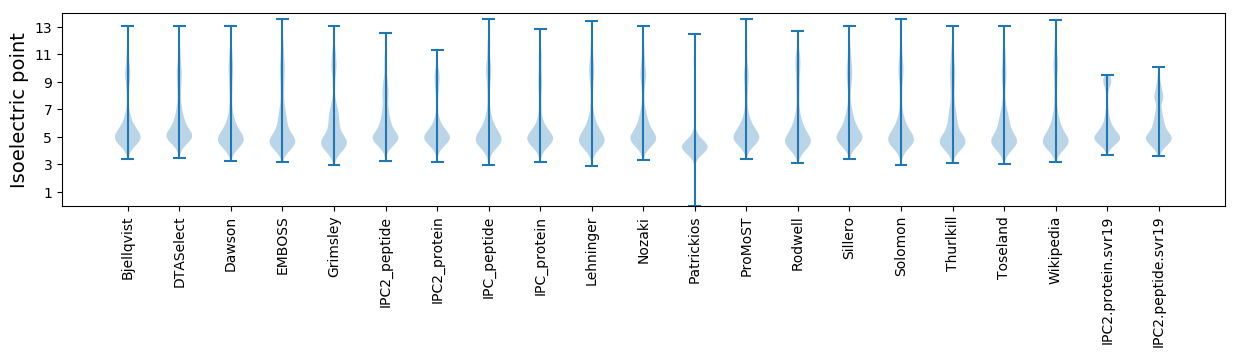
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z3YNI0|A0A2Z3YNI0_9CORY Putative N-succinyldiaminopimelate aminotransferase DapC OS=Corynebacterium provencense OX=1737425 GN=dapC PE=4 SV=1
MM1 pKa = 7.62PRR3 pKa = 11.84RR4 pKa = 11.84SAVPTTAMSMTAVVLALLTGCSGEE28 pKa = 4.91DD29 pKa = 3.38PAASPTFTAAEE40 pKa = 4.33TTSDD44 pKa = 3.39NTGGDD49 pKa = 3.19TGGWDD54 pKa = 3.29STGPQPSATSDD65 pKa = 3.2TVAADD70 pKa = 3.61AVSGTTTVTPQSGADD85 pKa = 3.61APSSVPGADD94 pKa = 3.18RR95 pKa = 11.84PASLTEE101 pKa = 4.31SGWSGSSVRR110 pKa = 11.84CNTGDD115 pKa = 3.12PWVYY119 pKa = 10.36AAEE122 pKa = 4.44GAPGQVVICLDD133 pKa = 4.07DD134 pKa = 4.62DD135 pKa = 3.81ALYY138 pKa = 10.72AVDD141 pKa = 3.99TDD143 pKa = 3.65NPYY146 pKa = 11.24GGDD149 pKa = 3.22NAYY152 pKa = 10.36AATSAVCPVSGTGRR166 pKa = 11.84DD167 pKa = 3.19RR168 pKa = 11.84YY169 pKa = 9.42VWHH172 pKa = 6.94HH173 pKa = 7.15ADD175 pKa = 3.83TGWTEE180 pKa = 3.83YY181 pKa = 11.05DD182 pKa = 3.0GTTRR186 pKa = 11.84YY187 pKa = 10.79VMDD190 pKa = 3.91IFSGRR195 pKa = 11.84GRR197 pKa = 11.84PDD199 pKa = 2.75LTDD202 pKa = 3.28AANIVEE208 pKa = 4.35DD209 pKa = 4.95AYY211 pKa = 11.19DD212 pKa = 4.1GAWTNPDD219 pKa = 3.19VPVPTLRR226 pKa = 11.84WCRR229 pKa = 11.84HH230 pKa = 5.73AGDD233 pKa = 4.21SPSTNLLAGG242 pKa = 3.81
MM1 pKa = 7.62PRR3 pKa = 11.84RR4 pKa = 11.84SAVPTTAMSMTAVVLALLTGCSGEE28 pKa = 4.91DD29 pKa = 3.38PAASPTFTAAEE40 pKa = 4.33TTSDD44 pKa = 3.39NTGGDD49 pKa = 3.19TGGWDD54 pKa = 3.29STGPQPSATSDD65 pKa = 3.2TVAADD70 pKa = 3.61AVSGTTTVTPQSGADD85 pKa = 3.61APSSVPGADD94 pKa = 3.18RR95 pKa = 11.84PASLTEE101 pKa = 4.31SGWSGSSVRR110 pKa = 11.84CNTGDD115 pKa = 3.12PWVYY119 pKa = 10.36AAEE122 pKa = 4.44GAPGQVVICLDD133 pKa = 4.07DD134 pKa = 4.62DD135 pKa = 3.81ALYY138 pKa = 10.72AVDD141 pKa = 3.99TDD143 pKa = 3.65NPYY146 pKa = 11.24GGDD149 pKa = 3.22NAYY152 pKa = 10.36AATSAVCPVSGTGRR166 pKa = 11.84DD167 pKa = 3.19RR168 pKa = 11.84YY169 pKa = 9.42VWHH172 pKa = 6.94HH173 pKa = 7.15ADD175 pKa = 3.83TGWTEE180 pKa = 3.83YY181 pKa = 11.05DD182 pKa = 3.0GTTRR186 pKa = 11.84YY187 pKa = 10.79VMDD190 pKa = 3.91IFSGRR195 pKa = 11.84GRR197 pKa = 11.84PDD199 pKa = 2.75LTDD202 pKa = 3.28AANIVEE208 pKa = 4.35DD209 pKa = 4.95AYY211 pKa = 11.19DD212 pKa = 4.1GAWTNPDD219 pKa = 3.19VPVPTLRR226 pKa = 11.84WCRR229 pKa = 11.84HH230 pKa = 5.73AGDD233 pKa = 4.21SPSTNLLAGG242 pKa = 3.81
Molecular weight: 24.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z3YLS0|A0A2Z3YLS0_9CORY Catalase OS=Corynebacterium provencense OX=1737425 GN=katA PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AHH17 pKa = 5.01KK18 pKa = 10.01HH19 pKa = 3.72GFRR22 pKa = 11.84TRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.67GRR42 pKa = 11.84KK43 pKa = 9.06SLTAA47 pKa = 4.07
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AHH17 pKa = 5.01KK18 pKa = 10.01HH19 pKa = 3.72GFRR22 pKa = 11.84TRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AIVSARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.67GRR42 pKa = 11.84KK43 pKa = 9.06SLTAA47 pKa = 4.07
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
922959 |
30 |
3136 |
346.8 |
37.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.902 ± 0.055 | 0.738 ± 0.014 |
6.709 ± 0.042 | 5.428 ± 0.042 |
2.921 ± 0.029 | 9.679 ± 0.045 |
2.068 ± 0.023 | 3.831 ± 0.034 |
1.971 ± 0.035 | 9.475 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.912 ± 0.021 | 2.119 ± 0.023 |
5.639 ± 0.036 | 2.567 ± 0.023 |
7.466 ± 0.051 | 5.986 ± 0.036 |
6.897 ± 0.039 | 9.357 ± 0.044 |
1.378 ± 0.019 | 1.957 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
