
Chondrocystis sp. NIES-4102
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Chroococcaceae; Chondrocystis; unclassified Chondrocystis
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
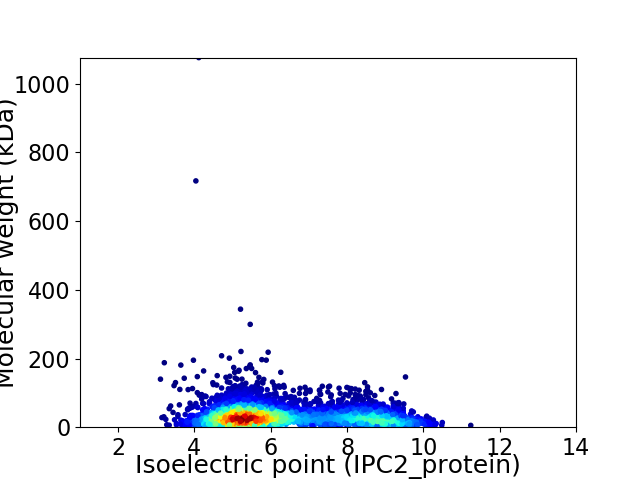
Virtual 2D-PAGE plot for 4342 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z4RSU1|A0A1Z4RSU1_9CHRO Putative ABC transporter permease protein OS=Chondrocystis sp. NIES-4102 OX=2005460 GN=NIES4102_25340 PE=3 SV=1
MM1 pKa = 7.58EE2 pKa = 5.89LFSNNNLIRR11 pKa = 11.84THH13 pKa = 7.07FLLQSEE19 pKa = 4.81WEE21 pKa = 3.94QGFTGKK27 pKa = 10.78LEE29 pKa = 4.04LTNTGEE35 pKa = 4.37TLKK38 pKa = 10.75DD39 pKa = 3.16WQIEE43 pKa = 4.4FEE45 pKa = 4.51SSWEE49 pKa = 3.88ITPGMIWGAEE59 pKa = 4.05IVSHH63 pKa = 6.13QGNKK67 pKa = 9.75YY68 pKa = 7.91VLKK71 pKa = 10.28PVDD74 pKa = 4.06YY75 pKa = 10.71NATIYY80 pKa = 10.38SQQQIAIIFNANKK93 pKa = 9.71IDD95 pKa = 3.81GQVGNPTNIIFSDD108 pKa = 4.32DD109 pKa = 3.74YY110 pKa = 11.74GLPTPPSNSSDD121 pKa = 3.2NSNSEE126 pKa = 4.45VINTNDD132 pKa = 4.99DD133 pKa = 3.6LATDD137 pKa = 3.5IDD139 pKa = 4.03FTLVKK144 pKa = 10.31DD145 pKa = 3.39WGSGFEE151 pKa = 4.15GQISITNNSPYY162 pKa = 10.75SIDD165 pKa = 3.15SWTLAFDD172 pKa = 4.43FPNPINNIWDD182 pKa = 3.73AEE184 pKa = 4.31IEE186 pKa = 4.28SSTNGSYY193 pKa = 9.92TIKK196 pKa = 10.04NAAWNRR202 pKa = 11.84EE203 pKa = 3.78LSAGEE208 pKa = 4.15TITFGFTGYY217 pKa = 11.01NSVTSQPQNFDD228 pKa = 3.51LDD230 pKa = 3.75GSTFASPSTSDD241 pKa = 2.93SKK243 pKa = 11.59YY244 pKa = 10.63SYY246 pKa = 11.16SNPDD250 pKa = 2.92LTTEE254 pKa = 4.46LEE256 pKa = 4.25LNKK259 pKa = 10.21NYY261 pKa = 10.1QGRR264 pKa = 11.84ATFYY268 pKa = 11.01DD269 pKa = 3.46AANPSGGIGASGYY282 pKa = 10.18DD283 pKa = 3.86VPSLDD288 pKa = 3.38QLYY291 pKa = 10.4KK292 pKa = 10.68VVAINNIQWNGSEE305 pKa = 3.8ASGGFFEE312 pKa = 5.63VSGPKK317 pKa = 9.39QRR319 pKa = 11.84EE320 pKa = 3.63GAAPVIVQVIDD331 pKa = 3.56YY332 pKa = 9.92LYY334 pKa = 10.72EE335 pKa = 4.31RR336 pKa = 11.84ADD338 pKa = 3.66GMDD341 pKa = 3.34MSAEE345 pKa = 3.96AFKK348 pKa = 11.09LVADD352 pKa = 4.18PVDD355 pKa = 4.37GIVNINYY362 pKa = 9.66QLVGPDD368 pKa = 3.73DD369 pKa = 5.56DD370 pKa = 5.02YY371 pKa = 11.64VTAYY375 pKa = 10.07GYY377 pKa = 10.81SIGQGIVVEE386 pKa = 6.01GIAEE390 pKa = 4.26SNPYY394 pKa = 8.53YY395 pKa = 10.85AAVRR399 pKa = 11.84LNNYY403 pKa = 8.62RR404 pKa = 11.84YY405 pKa = 9.49PIEE408 pKa = 4.14TVEE411 pKa = 4.48LLTEE415 pKa = 4.09NGEE418 pKa = 4.51AIDD421 pKa = 5.26LNRR424 pKa = 11.84EE425 pKa = 3.62SDD427 pKa = 3.45NRR429 pKa = 11.84FVLDD433 pKa = 3.9GNYY436 pKa = 9.92PLYY439 pKa = 10.6GAQDD443 pKa = 4.12LLVTDD448 pKa = 4.2IFGQQVTLDD457 pKa = 4.22DD458 pKa = 4.08VNITNGSSNDD468 pKa = 3.68TVTGEE473 pKa = 4.04QFAKK477 pKa = 10.41IAA479 pKa = 3.88
MM1 pKa = 7.58EE2 pKa = 5.89LFSNNNLIRR11 pKa = 11.84THH13 pKa = 7.07FLLQSEE19 pKa = 4.81WEE21 pKa = 3.94QGFTGKK27 pKa = 10.78LEE29 pKa = 4.04LTNTGEE35 pKa = 4.37TLKK38 pKa = 10.75DD39 pKa = 3.16WQIEE43 pKa = 4.4FEE45 pKa = 4.51SSWEE49 pKa = 3.88ITPGMIWGAEE59 pKa = 4.05IVSHH63 pKa = 6.13QGNKK67 pKa = 9.75YY68 pKa = 7.91VLKK71 pKa = 10.28PVDD74 pKa = 4.06YY75 pKa = 10.71NATIYY80 pKa = 10.38SQQQIAIIFNANKK93 pKa = 9.71IDD95 pKa = 3.81GQVGNPTNIIFSDD108 pKa = 4.32DD109 pKa = 3.74YY110 pKa = 11.74GLPTPPSNSSDD121 pKa = 3.2NSNSEE126 pKa = 4.45VINTNDD132 pKa = 4.99DD133 pKa = 3.6LATDD137 pKa = 3.5IDD139 pKa = 4.03FTLVKK144 pKa = 10.31DD145 pKa = 3.39WGSGFEE151 pKa = 4.15GQISITNNSPYY162 pKa = 10.75SIDD165 pKa = 3.15SWTLAFDD172 pKa = 4.43FPNPINNIWDD182 pKa = 3.73AEE184 pKa = 4.31IEE186 pKa = 4.28SSTNGSYY193 pKa = 9.92TIKK196 pKa = 10.04NAAWNRR202 pKa = 11.84EE203 pKa = 3.78LSAGEE208 pKa = 4.15TITFGFTGYY217 pKa = 11.01NSVTSQPQNFDD228 pKa = 3.51LDD230 pKa = 3.75GSTFASPSTSDD241 pKa = 2.93SKK243 pKa = 11.59YY244 pKa = 10.63SYY246 pKa = 11.16SNPDD250 pKa = 2.92LTTEE254 pKa = 4.46LEE256 pKa = 4.25LNKK259 pKa = 10.21NYY261 pKa = 10.1QGRR264 pKa = 11.84ATFYY268 pKa = 11.01DD269 pKa = 3.46AANPSGGIGASGYY282 pKa = 10.18DD283 pKa = 3.86VPSLDD288 pKa = 3.38QLYY291 pKa = 10.4KK292 pKa = 10.68VVAINNIQWNGSEE305 pKa = 3.8ASGGFFEE312 pKa = 5.63VSGPKK317 pKa = 9.39QRR319 pKa = 11.84EE320 pKa = 3.63GAAPVIVQVIDD331 pKa = 3.56YY332 pKa = 9.92LYY334 pKa = 10.72EE335 pKa = 4.31RR336 pKa = 11.84ADD338 pKa = 3.66GMDD341 pKa = 3.34MSAEE345 pKa = 3.96AFKK348 pKa = 11.09LVADD352 pKa = 4.18PVDD355 pKa = 4.37GIVNINYY362 pKa = 9.66QLVGPDD368 pKa = 3.73DD369 pKa = 5.56DD370 pKa = 5.02YY371 pKa = 11.64VTAYY375 pKa = 10.07GYY377 pKa = 10.81SIGQGIVVEE386 pKa = 6.01GIAEE390 pKa = 4.26SNPYY394 pKa = 8.53YY395 pKa = 10.85AAVRR399 pKa = 11.84LNNYY403 pKa = 8.62RR404 pKa = 11.84YY405 pKa = 9.49PIEE408 pKa = 4.14TVEE411 pKa = 4.48LLTEE415 pKa = 4.09NGEE418 pKa = 4.51AIDD421 pKa = 5.26LNRR424 pKa = 11.84EE425 pKa = 3.62SDD427 pKa = 3.45NRR429 pKa = 11.84FVLDD433 pKa = 3.9GNYY436 pKa = 9.92PLYY439 pKa = 10.6GAQDD443 pKa = 4.12LLVTDD448 pKa = 4.2IFGQQVTLDD457 pKa = 4.22DD458 pKa = 4.08VNITNGSSNDD468 pKa = 3.68TVTGEE473 pKa = 4.04QFAKK477 pKa = 10.41IAA479 pKa = 3.88
Molecular weight: 52.7 kDa
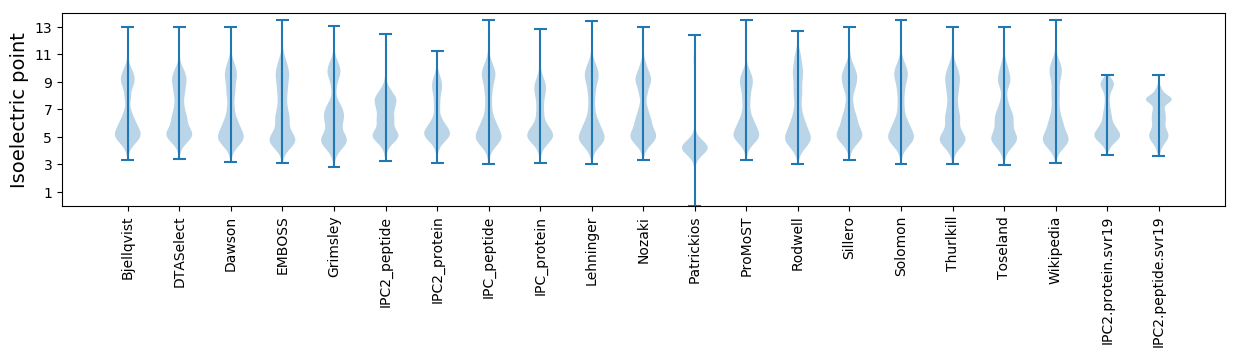
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z4RKU0|A0A1Z4RKU0_9CHRO Uncharacterized protein OS=Chondrocystis sp. NIES-4102 OX=2005460 GN=NIES4102_00810 PE=4 SV=1
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TSNGKK29 pKa = 9.45RR30 pKa = 11.84VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.07KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
MM1 pKa = 6.61TQRR4 pKa = 11.84TLGGTNRR11 pKa = 11.84KK12 pKa = 7.61QKK14 pKa = 8.99RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TSNGKK29 pKa = 9.45RR30 pKa = 11.84VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.07KK38 pKa = 9.65GRR40 pKa = 11.84HH41 pKa = 5.0RR42 pKa = 11.84LSVV45 pKa = 3.12
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1365604 |
29 |
10077 |
314.5 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.685 ± 0.039 | 0.984 ± 0.018 |
5.074 ± 0.043 | 6.071 ± 0.039 |
3.82 ± 0.023 | 6.308 ± 0.048 |
1.698 ± 0.021 | 7.905 ± 0.034 |
5.444 ± 0.04 | 10.963 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.021 | 5.124 ± 0.047 |
4.24 ± 0.03 | 5.565 ± 0.039 |
4.428 ± 0.034 | 6.463 ± 0.031 |
5.711 ± 0.042 | 6.078 ± 0.03 |
1.357 ± 0.017 | 3.321 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
