
Nocardioides sp. Root614
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
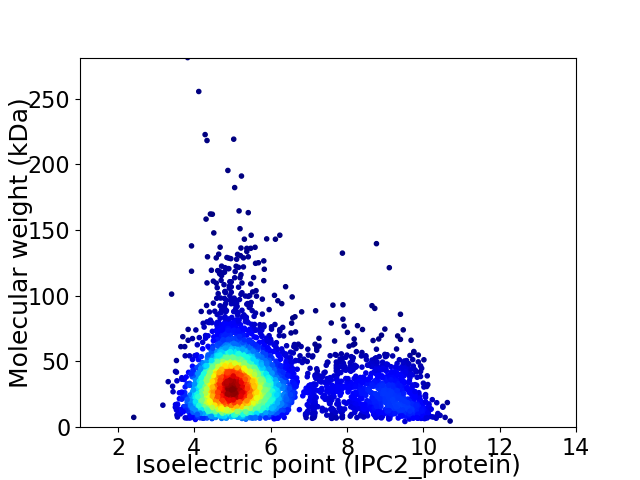
Virtual 2D-PAGE plot for 4780 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q8DUZ2|A0A0Q8DUZ2_9ACTN Multidrug MFS transporter OS=Nocardioides sp. Root614 OX=1736571 GN=ASD81_15580 PE=4 SV=1
MM1 pKa = 7.33TPASLHH7 pKa = 5.7LFSRR11 pKa = 11.84DD12 pKa = 3.13RR13 pKa = 11.84LATRR17 pKa = 11.84VTAVGLFACGSWALAFSATAAAPASADD44 pKa = 3.64AGDD47 pKa = 4.05LTCHH51 pKa = 6.15GKK53 pKa = 10.22AVTILVDD60 pKa = 3.93ASHH63 pKa = 6.75NGSPVYY69 pKa = 10.43GSSGDD74 pKa = 3.8DD75 pKa = 3.21VIMAFGGGHH84 pKa = 6.92HH85 pKa = 7.37IDD87 pKa = 3.59GQAGNDD93 pKa = 4.17TICASSDD100 pKa = 3.3VGSVGDD106 pKa = 4.05VILGGVGNDD115 pKa = 3.66YY116 pKa = 9.95LTGGAGNDD124 pKa = 3.8TLDD127 pKa = 4.09GGSDD131 pKa = 3.13QDD133 pKa = 4.76FIGGGPGDD141 pKa = 4.5DD142 pKa = 3.57NLFGGDD148 pKa = 3.8HH149 pKa = 6.15VDD151 pKa = 3.46EE152 pKa = 4.76LRR154 pKa = 11.84GGSGDD159 pKa = 4.05DD160 pKa = 3.39RR161 pKa = 11.84LTGGDD166 pKa = 4.33ARR168 pKa = 11.84DD169 pKa = 3.68MLYY172 pKa = 10.9GDD174 pKa = 4.41SGDD177 pKa = 5.06DD178 pKa = 3.72VIHH181 pKa = 6.81GNDD184 pKa = 4.1GGDD187 pKa = 3.21GVYY190 pKa = 10.82GQDD193 pKa = 5.06GNDD196 pKa = 3.42HH197 pKa = 7.07LYY199 pKa = 11.38GEE201 pKa = 4.92IGQDD205 pKa = 3.42VLVGGLGDD213 pKa = 4.5DD214 pKa = 4.16LLHH217 pKa = 6.87GGLYY221 pKa = 9.9PDD223 pKa = 3.86SLYY226 pKa = 11.03GQEE229 pKa = 4.51GDD231 pKa = 3.77DD232 pKa = 4.75VIYY235 pKa = 10.5TGDD238 pKa = 3.72GGPAGFDD245 pKa = 3.2KK246 pKa = 11.36AFGDD250 pKa = 3.74SGNDD254 pKa = 3.44VLHH257 pKa = 6.73GSSDD261 pKa = 3.71GDD263 pKa = 3.57RR264 pKa = 11.84LNGGTGNDD272 pKa = 3.54LLYY275 pKa = 10.84GYY277 pKa = 10.51GSQDD281 pKa = 2.95KK282 pKa = 10.93LYY284 pKa = 10.63GDD286 pKa = 4.19SGNDD290 pKa = 3.14RR291 pKa = 11.84LEE293 pKa = 4.4GGGNRR298 pKa = 11.84DD299 pKa = 3.4EE300 pKa = 4.93LRR302 pKa = 11.84GHH304 pKa = 6.72EE305 pKa = 4.53GNDD308 pKa = 3.01RR309 pKa = 11.84LYY311 pKa = 11.2GGDD314 pKa = 4.03GADD317 pKa = 4.13DD318 pKa = 3.56LHH320 pKa = 8.69GGVNDD325 pKa = 4.75DD326 pKa = 3.99YY327 pKa = 11.9LNGGGDD333 pKa = 5.74LMDD336 pKa = 4.82QGNGDD341 pKa = 4.32PGVDD345 pKa = 3.11TCVGIRR351 pKa = 11.84DD352 pKa = 3.87AMQTTCEE359 pKa = 3.9RR360 pKa = 3.79
MM1 pKa = 7.33TPASLHH7 pKa = 5.7LFSRR11 pKa = 11.84DD12 pKa = 3.13RR13 pKa = 11.84LATRR17 pKa = 11.84VTAVGLFACGSWALAFSATAAAPASADD44 pKa = 3.64AGDD47 pKa = 4.05LTCHH51 pKa = 6.15GKK53 pKa = 10.22AVTILVDD60 pKa = 3.93ASHH63 pKa = 6.75NGSPVYY69 pKa = 10.43GSSGDD74 pKa = 3.8DD75 pKa = 3.21VIMAFGGGHH84 pKa = 6.92HH85 pKa = 7.37IDD87 pKa = 3.59GQAGNDD93 pKa = 4.17TICASSDD100 pKa = 3.3VGSVGDD106 pKa = 4.05VILGGVGNDD115 pKa = 3.66YY116 pKa = 9.95LTGGAGNDD124 pKa = 3.8TLDD127 pKa = 4.09GGSDD131 pKa = 3.13QDD133 pKa = 4.76FIGGGPGDD141 pKa = 4.5DD142 pKa = 3.57NLFGGDD148 pKa = 3.8HH149 pKa = 6.15VDD151 pKa = 3.46EE152 pKa = 4.76LRR154 pKa = 11.84GGSGDD159 pKa = 4.05DD160 pKa = 3.39RR161 pKa = 11.84LTGGDD166 pKa = 4.33ARR168 pKa = 11.84DD169 pKa = 3.68MLYY172 pKa = 10.9GDD174 pKa = 4.41SGDD177 pKa = 5.06DD178 pKa = 3.72VIHH181 pKa = 6.81GNDD184 pKa = 4.1GGDD187 pKa = 3.21GVYY190 pKa = 10.82GQDD193 pKa = 5.06GNDD196 pKa = 3.42HH197 pKa = 7.07LYY199 pKa = 11.38GEE201 pKa = 4.92IGQDD205 pKa = 3.42VLVGGLGDD213 pKa = 4.5DD214 pKa = 4.16LLHH217 pKa = 6.87GGLYY221 pKa = 9.9PDD223 pKa = 3.86SLYY226 pKa = 11.03GQEE229 pKa = 4.51GDD231 pKa = 3.77DD232 pKa = 4.75VIYY235 pKa = 10.5TGDD238 pKa = 3.72GGPAGFDD245 pKa = 3.2KK246 pKa = 11.36AFGDD250 pKa = 3.74SGNDD254 pKa = 3.44VLHH257 pKa = 6.73GSSDD261 pKa = 3.71GDD263 pKa = 3.57RR264 pKa = 11.84LNGGTGNDD272 pKa = 3.54LLYY275 pKa = 10.84GYY277 pKa = 10.51GSQDD281 pKa = 2.95KK282 pKa = 10.93LYY284 pKa = 10.63GDD286 pKa = 4.19SGNDD290 pKa = 3.14RR291 pKa = 11.84LEE293 pKa = 4.4GGGNRR298 pKa = 11.84DD299 pKa = 3.4EE300 pKa = 4.93LRR302 pKa = 11.84GHH304 pKa = 6.72EE305 pKa = 4.53GNDD308 pKa = 3.01RR309 pKa = 11.84LYY311 pKa = 11.2GGDD314 pKa = 4.03GADD317 pKa = 4.13DD318 pKa = 3.56LHH320 pKa = 8.69GGVNDD325 pKa = 4.75DD326 pKa = 3.99YY327 pKa = 11.9LNGGGDD333 pKa = 5.74LMDD336 pKa = 4.82QGNGDD341 pKa = 4.32PGVDD345 pKa = 3.11TCVGIRR351 pKa = 11.84DD352 pKa = 3.87AMQTTCEE359 pKa = 3.9RR360 pKa = 3.79
Molecular weight: 36.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q8D7T6|A0A0Q8D7T6_9ACTN Uncharacterized protein OS=Nocardioides sp. Root614 OX=1736571 GN=ASD81_24350 PE=4 SV=1
MM1 pKa = 7.64RR2 pKa = 11.84PVSGIGSRR10 pKa = 11.84RR11 pKa = 11.84SVEE14 pKa = 3.4QWDD17 pKa = 3.74RR18 pKa = 11.84FFEE21 pKa = 4.36LVCAGASVSAAAAEE35 pKa = 4.17IGVARR40 pKa = 11.84QSAWEE45 pKa = 4.0RR46 pKa = 11.84WRR48 pKa = 11.84NSAPMEE54 pKa = 4.27LKK56 pKa = 10.57LLQGRR61 pKa = 11.84GGGLGQPAEE70 pKa = 4.49CEE72 pKa = 3.99QVAGRR77 pKa = 11.84VRR79 pKa = 11.84RR80 pKa = 11.84ALTSEE85 pKa = 3.7DD86 pKa = 3.33RR87 pKa = 11.84AVIAAGVRR95 pKa = 11.84KK96 pKa = 9.26KK97 pKa = 7.42WTYY100 pKa = 11.46AEE102 pKa = 3.87IGEE105 pKa = 4.61AIGRR109 pKa = 11.84SPSVICRR116 pKa = 11.84EE117 pKa = 3.94VARR120 pKa = 11.84NKK122 pKa = 10.72GPDD125 pKa = 3.41GSYY128 pKa = 10.5HH129 pKa = 5.55GTLAHH134 pKa = 6.62AAARR138 pKa = 11.84QRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84PKK145 pKa = 9.45EE146 pKa = 3.46FRR148 pKa = 11.84LVAEE152 pKa = 4.83PGLCRR157 pKa = 11.84RR158 pKa = 11.84IEE160 pKa = 3.74TWMDD164 pKa = 3.31EE165 pKa = 3.91GWSPKK170 pKa = 10.34LIARR174 pKa = 11.84VLRR177 pKa = 11.84EE178 pKa = 3.81EE179 pKa = 4.28SPRR182 pKa = 11.84MIMGRR187 pKa = 11.84VSHH190 pKa = 5.25EE191 pKa = 4.14TIYY194 pKa = 10.62QALYY198 pKa = 8.06VQTRR202 pKa = 11.84GSLRR206 pKa = 11.84KK207 pKa = 9.51DD208 pKa = 3.92LYY210 pKa = 10.57RR211 pKa = 11.84QLSTTRR217 pKa = 11.84PRR219 pKa = 11.84RR220 pKa = 11.84QPRR223 pKa = 11.84VGAKK227 pKa = 8.19RR228 pKa = 11.84TNSPYY233 pKa = 10.62KK234 pKa = 9.66EE235 pKa = 3.99AFKK238 pKa = 10.55ISQRR242 pKa = 11.84PAEE245 pKa = 4.27VADD248 pKa = 4.12RR249 pKa = 11.84AVPGHH254 pKa = 6.24WEE256 pKa = 3.87GDD258 pKa = 3.61LVMGTAGGVAVGTLVEE274 pKa = 4.19RR275 pKa = 11.84STRR278 pKa = 11.84FTILLHH284 pKa = 6.34LPGRR288 pKa = 11.84HH289 pKa = 5.98DD290 pKa = 4.16AEE292 pKa = 4.37SVAAAMIRR300 pKa = 11.84EE301 pKa = 4.11MSEE304 pKa = 3.95LPEE307 pKa = 4.33HH308 pKa = 6.71LRR310 pKa = 11.84QSLTWDD316 pKa = 3.27RR317 pKa = 11.84GTEE320 pKa = 3.67LARR323 pKa = 11.84YY324 pKa = 9.58DD325 pKa = 5.12KK326 pKa = 10.86IQTDD330 pKa = 3.85LAMPVYY336 pKa = 10.35FCDD339 pKa = 4.35PHH341 pKa = 8.5APWQRR346 pKa = 11.84GTNEE350 pKa = 3.5NTNRR354 pKa = 11.84LLRR357 pKa = 11.84HH358 pKa = 6.0WLTKK362 pKa = 9.89GTDD365 pKa = 3.39LSRR368 pKa = 11.84FSAADD373 pKa = 3.35LRR375 pKa = 11.84RR376 pKa = 11.84IAASLNARR384 pKa = 11.84PRR386 pKa = 11.84PTLNMQTPAQALDD399 pKa = 3.53QLLTNPAAAA408 pKa = 4.6
MM1 pKa = 7.64RR2 pKa = 11.84PVSGIGSRR10 pKa = 11.84RR11 pKa = 11.84SVEE14 pKa = 3.4QWDD17 pKa = 3.74RR18 pKa = 11.84FFEE21 pKa = 4.36LVCAGASVSAAAAEE35 pKa = 4.17IGVARR40 pKa = 11.84QSAWEE45 pKa = 4.0RR46 pKa = 11.84WRR48 pKa = 11.84NSAPMEE54 pKa = 4.27LKK56 pKa = 10.57LLQGRR61 pKa = 11.84GGGLGQPAEE70 pKa = 4.49CEE72 pKa = 3.99QVAGRR77 pKa = 11.84VRR79 pKa = 11.84RR80 pKa = 11.84ALTSEE85 pKa = 3.7DD86 pKa = 3.33RR87 pKa = 11.84AVIAAGVRR95 pKa = 11.84KK96 pKa = 9.26KK97 pKa = 7.42WTYY100 pKa = 11.46AEE102 pKa = 3.87IGEE105 pKa = 4.61AIGRR109 pKa = 11.84SPSVICRR116 pKa = 11.84EE117 pKa = 3.94VARR120 pKa = 11.84NKK122 pKa = 10.72GPDD125 pKa = 3.41GSYY128 pKa = 10.5HH129 pKa = 5.55GTLAHH134 pKa = 6.62AAARR138 pKa = 11.84QRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 11.84PKK145 pKa = 9.45EE146 pKa = 3.46FRR148 pKa = 11.84LVAEE152 pKa = 4.83PGLCRR157 pKa = 11.84RR158 pKa = 11.84IEE160 pKa = 3.74TWMDD164 pKa = 3.31EE165 pKa = 3.91GWSPKK170 pKa = 10.34LIARR174 pKa = 11.84VLRR177 pKa = 11.84EE178 pKa = 3.81EE179 pKa = 4.28SPRR182 pKa = 11.84MIMGRR187 pKa = 11.84VSHH190 pKa = 5.25EE191 pKa = 4.14TIYY194 pKa = 10.62QALYY198 pKa = 8.06VQTRR202 pKa = 11.84GSLRR206 pKa = 11.84KK207 pKa = 9.51DD208 pKa = 3.92LYY210 pKa = 10.57RR211 pKa = 11.84QLSTTRR217 pKa = 11.84PRR219 pKa = 11.84RR220 pKa = 11.84QPRR223 pKa = 11.84VGAKK227 pKa = 8.19RR228 pKa = 11.84TNSPYY233 pKa = 10.62KK234 pKa = 9.66EE235 pKa = 3.99AFKK238 pKa = 10.55ISQRR242 pKa = 11.84PAEE245 pKa = 4.27VADD248 pKa = 4.12RR249 pKa = 11.84AVPGHH254 pKa = 6.24WEE256 pKa = 3.87GDD258 pKa = 3.61LVMGTAGGVAVGTLVEE274 pKa = 4.19RR275 pKa = 11.84STRR278 pKa = 11.84FTILLHH284 pKa = 6.34LPGRR288 pKa = 11.84HH289 pKa = 5.98DD290 pKa = 4.16AEE292 pKa = 4.37SVAAAMIRR300 pKa = 11.84EE301 pKa = 4.11MSEE304 pKa = 3.95LPEE307 pKa = 4.33HH308 pKa = 6.71LRR310 pKa = 11.84QSLTWDD316 pKa = 3.27RR317 pKa = 11.84GTEE320 pKa = 3.67LARR323 pKa = 11.84YY324 pKa = 9.58DD325 pKa = 5.12KK326 pKa = 10.86IQTDD330 pKa = 3.85LAMPVYY336 pKa = 10.35FCDD339 pKa = 4.35PHH341 pKa = 8.5APWQRR346 pKa = 11.84GTNEE350 pKa = 3.5NTNRR354 pKa = 11.84LLRR357 pKa = 11.84HH358 pKa = 6.0WLTKK362 pKa = 9.89GTDD365 pKa = 3.39LSRR368 pKa = 11.84FSAADD373 pKa = 3.35LRR375 pKa = 11.84RR376 pKa = 11.84IAASLNARR384 pKa = 11.84PRR386 pKa = 11.84PTLNMQTPAQALDD399 pKa = 3.53QLLTNPAAAA408 pKa = 4.6
Molecular weight: 45.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1577883 |
37 |
2876 |
330.1 |
35.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.864 ± 0.048 | 0.747 ± 0.01 |
6.605 ± 0.034 | 5.647 ± 0.034 |
2.923 ± 0.02 | 9.133 ± 0.032 |
2.121 ± 0.018 | 3.947 ± 0.022 |
2.272 ± 0.027 | 10.059 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.81 ± 0.013 | 2.066 ± 0.022 |
5.491 ± 0.027 | 2.816 ± 0.018 |
7.106 ± 0.042 | 5.449 ± 0.024 |
6.262 ± 0.041 | 9.191 ± 0.033 |
1.49 ± 0.014 | 2.0 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
