
Aspergillus sclerotioniger CBS 115572
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus sclerotioniger
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
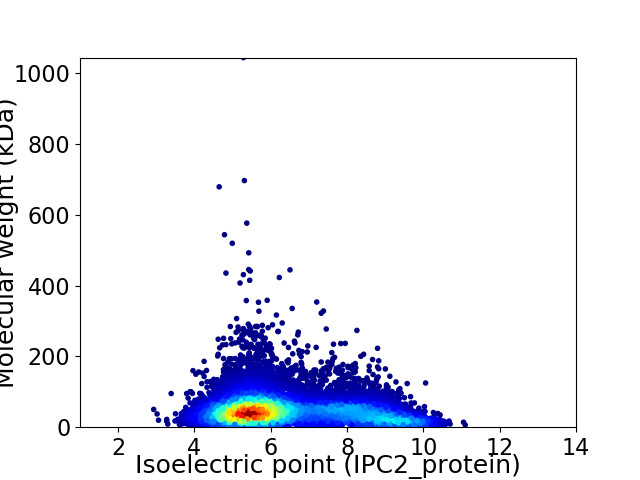
Virtual 2D-PAGE plot for 12304 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317WSN0|A0A317WSN0_9EURO DNA mismatch repair protein Msh2 OS=Aspergillus sclerotioniger CBS 115572 OX=1450535 GN=BO94DRAFT_534891 PE=3 SV=1
MM1 pKa = 6.5VTSSSLIAVGLTLWASLVSATPADD25 pKa = 3.91PRR27 pKa = 11.84VTAAPKK33 pKa = 10.88LEE35 pKa = 4.8DD36 pKa = 3.6LDD38 pKa = 4.91KK39 pKa = 11.08RR40 pKa = 11.84ATSCTFSGSEE50 pKa = 4.27GASSASKK57 pKa = 10.82SKK59 pKa = 9.53TSCSTIVLSDD69 pKa = 3.44VAVPSGTTLDD79 pKa = 3.71LTDD82 pKa = 4.74LNEE85 pKa = 4.24GTHH88 pKa = 6.42VIFEE92 pKa = 4.49GEE94 pKa = 4.12TTFGYY99 pKa = 10.09EE100 pKa = 3.78EE101 pKa = 4.23WDD103 pKa = 3.84GPLVSVSGTDD113 pKa = 3.23ITVTGADD120 pKa = 3.38GAYY123 pKa = 10.71LNGDD127 pKa = 3.92GSRR130 pKa = 11.84WWDD133 pKa = 3.38DD134 pKa = 3.26EE135 pKa = 4.13GSNGGKK141 pKa = 8.24TKK143 pKa = 10.59PKK145 pKa = 10.15FFYY148 pKa = 9.6AHH150 pKa = 6.08NMIDD154 pKa = 3.63STISGIYY161 pKa = 8.6IQNSPVQVFSIDD173 pKa = 3.43DD174 pKa = 3.53SSYY177 pKa = 10.58LTLEE181 pKa = 5.51DD182 pKa = 3.4ITIDD186 pKa = 5.33DD187 pKa = 4.51SDD189 pKa = 5.3GDD191 pKa = 4.01DD192 pKa = 3.86AGAANTDD199 pKa = 3.24GFDD202 pKa = 3.65IGSSTYY208 pKa = 9.33ITITGANVYY217 pKa = 10.14NQDD220 pKa = 3.1DD221 pKa = 4.24CVAVNSGEE229 pKa = 4.07NIYY232 pKa = 10.7FSGGVCSGGHH242 pKa = 5.7GLSIGSVGGRR252 pKa = 11.84DD253 pKa = 3.68DD254 pKa = 3.79NTVKK258 pKa = 10.8NVTFYY263 pKa = 11.21NSEE266 pKa = 3.87IKK268 pKa = 10.57SSQNGVRR275 pKa = 11.84IKK277 pKa = 9.88TIYY280 pKa = 10.53GDD282 pKa = 3.58TGSVSDD288 pKa = 3.59ITYY291 pKa = 10.76EE292 pKa = 4.34KK293 pKa = 9.38ITLSEE298 pKa = 3.69ITKK301 pKa = 9.75YY302 pKa = 10.99GIVVEE307 pKa = 4.11QNYY310 pKa = 10.92DD311 pKa = 3.58DD312 pKa = 4.16TSKK315 pKa = 11.27SPTDD319 pKa = 4.34GITIEE324 pKa = 4.51NFILDD329 pKa = 4.06GVEE332 pKa = 4.18GTVEE336 pKa = 3.89SSGTNIYY343 pKa = 9.42IVCGSDD349 pKa = 4.0SCTDD353 pKa = 3.53WTWTTVDD360 pKa = 2.81ITGGKK365 pKa = 7.95TSSDD369 pKa = 4.06CEE371 pKa = 4.14NVPDD375 pKa = 6.29DD376 pKa = 4.16ISCC379 pKa = 3.93
MM1 pKa = 6.5VTSSSLIAVGLTLWASLVSATPADD25 pKa = 3.91PRR27 pKa = 11.84VTAAPKK33 pKa = 10.88LEE35 pKa = 4.8DD36 pKa = 3.6LDD38 pKa = 4.91KK39 pKa = 11.08RR40 pKa = 11.84ATSCTFSGSEE50 pKa = 4.27GASSASKK57 pKa = 10.82SKK59 pKa = 9.53TSCSTIVLSDD69 pKa = 3.44VAVPSGTTLDD79 pKa = 3.71LTDD82 pKa = 4.74LNEE85 pKa = 4.24GTHH88 pKa = 6.42VIFEE92 pKa = 4.49GEE94 pKa = 4.12TTFGYY99 pKa = 10.09EE100 pKa = 3.78EE101 pKa = 4.23WDD103 pKa = 3.84GPLVSVSGTDD113 pKa = 3.23ITVTGADD120 pKa = 3.38GAYY123 pKa = 10.71LNGDD127 pKa = 3.92GSRR130 pKa = 11.84WWDD133 pKa = 3.38DD134 pKa = 3.26EE135 pKa = 4.13GSNGGKK141 pKa = 8.24TKK143 pKa = 10.59PKK145 pKa = 10.15FFYY148 pKa = 9.6AHH150 pKa = 6.08NMIDD154 pKa = 3.63STISGIYY161 pKa = 8.6IQNSPVQVFSIDD173 pKa = 3.43DD174 pKa = 3.53SSYY177 pKa = 10.58LTLEE181 pKa = 5.51DD182 pKa = 3.4ITIDD186 pKa = 5.33DD187 pKa = 4.51SDD189 pKa = 5.3GDD191 pKa = 4.01DD192 pKa = 3.86AGAANTDD199 pKa = 3.24GFDD202 pKa = 3.65IGSSTYY208 pKa = 9.33ITITGANVYY217 pKa = 10.14NQDD220 pKa = 3.1DD221 pKa = 4.24CVAVNSGEE229 pKa = 4.07NIYY232 pKa = 10.7FSGGVCSGGHH242 pKa = 5.7GLSIGSVGGRR252 pKa = 11.84DD253 pKa = 3.68DD254 pKa = 3.79NTVKK258 pKa = 10.8NVTFYY263 pKa = 11.21NSEE266 pKa = 3.87IKK268 pKa = 10.57SSQNGVRR275 pKa = 11.84IKK277 pKa = 9.88TIYY280 pKa = 10.53GDD282 pKa = 3.58TGSVSDD288 pKa = 3.59ITYY291 pKa = 10.76EE292 pKa = 4.34KK293 pKa = 9.38ITLSEE298 pKa = 3.69ITKK301 pKa = 9.75YY302 pKa = 10.99GIVVEE307 pKa = 4.11QNYY310 pKa = 10.92DD311 pKa = 3.58DD312 pKa = 4.16TSKK315 pKa = 11.27SPTDD319 pKa = 4.34GITIEE324 pKa = 4.51NFILDD329 pKa = 4.06GVEE332 pKa = 4.18GTVEE336 pKa = 3.89SSGTNIYY343 pKa = 9.42IVCGSDD349 pKa = 4.0SCTDD353 pKa = 3.53WTWTTVDD360 pKa = 2.81ITGGKK365 pKa = 7.95TSSDD369 pKa = 4.06CEE371 pKa = 4.14NVPDD375 pKa = 6.29DD376 pKa = 4.16ISCC379 pKa = 3.93
Molecular weight: 39.83 kDa
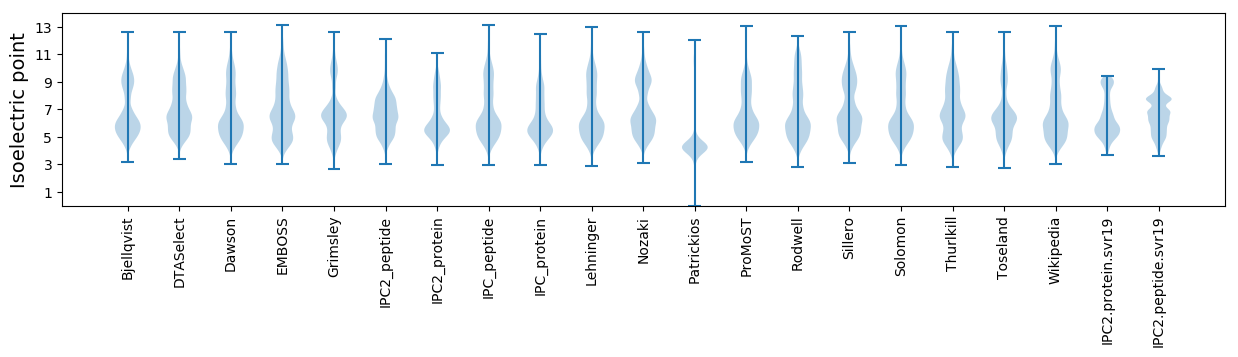
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317WJ69|A0A317WJ69_9EURO DUF1212-domain-containing protein OS=Aspergillus sclerotioniger CBS 115572 OX=1450535 GN=BO94DRAFT_467054 PE=3 SV=1
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.54LAKK15 pKa = 9.74AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
MM1 pKa = 7.88PSHH4 pKa = 6.91KK5 pKa = 10.39SFRR8 pKa = 11.84TKK10 pKa = 10.45QKK12 pKa = 9.54LAKK15 pKa = 9.74AQRR18 pKa = 11.84QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.14WRR45 pKa = 11.84KK46 pKa = 7.41TRR48 pKa = 11.84LGII51 pKa = 4.46
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5731707 |
49 |
9565 |
465.8 |
51.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.316 ± 0.02 | 1.334 ± 0.01 |
5.591 ± 0.016 | 6.008 ± 0.025 |
3.769 ± 0.013 | 6.814 ± 0.018 |
2.497 ± 0.01 | 5.037 ± 0.016 |
4.314 ± 0.02 | 9.258 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.2 ± 0.009 | 3.53 ± 0.011 |
6.094 ± 0.026 | 3.989 ± 0.016 |
6.159 ± 0.018 | 8.29 ± 0.024 |
6.071 ± 0.018 | 6.274 ± 0.019 |
1.536 ± 0.009 | 2.92 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
