
Olleya aquimaris
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Olleya
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
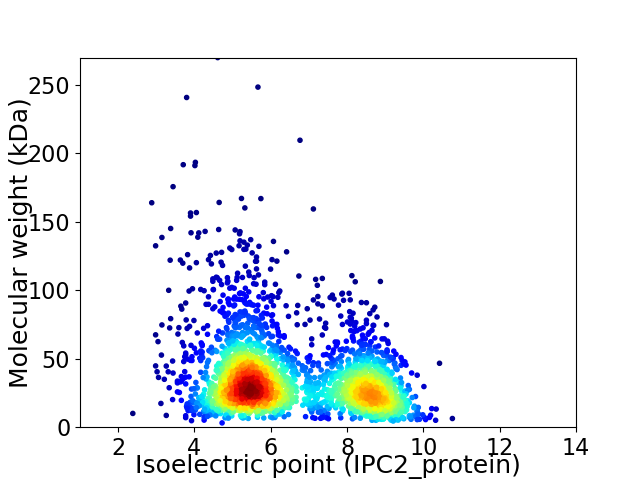
Virtual 2D-PAGE plot for 2642 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A327R5Q1|A0A327R5Q1_9FLAO Outer membrane receptor protein involved in Fe transport OS=Olleya aquimaris OX=639310 GN=LY08_02422 PE=3 SV=1
DDD2 pKa = 5.84IDDD5 pKa = 3.66NCDDD9 pKa = 3.07NIDDD13 pKa = 3.75EE14 pKa = 6.35FDDD17 pKa = 3.4DDD19 pKa = 3.97DDD21 pKa = 4.02FTACQGDDD29 pKa = 4.7DDD31 pKa = 5.45DD32 pKa = 5.3DDD34 pKa = 4.19NSYYY38 pKa = 10.12GAPEEE43 pKa = 3.83CDDD46 pKa = 5.83IDDD49 pKa = 3.54NCDDD53 pKa = 3.27LVDDD57 pKa = 4.18DDD59 pKa = 4.23DD60 pKa = 3.87SVIGLPDDD68 pKa = 3.3VITEEE73 pKa = 4.18PLGDDD78 pKa = 3.84CQGVAEEE85 pKa = 4.25TVDDD89 pKa = 3.53LNASDDD95 pKa = 4.74SSISYYY101 pKa = 9.43WSNGSTDDD109 pKa = 2.79SIIVSDDD116 pKa = 3.57NTYYY120 pKa = 8.97VSVTGNSGCTTTSSYYY136 pKa = 10.75STAVPSDDD144 pKa = 3.61LSGYYY149 pKa = 10.36MLANKKK155 pKa = 9.68EE156 pKa = 4.13YYY158 pKa = 10.47KKK160 pKa = 9.62EE161 pKa = 4.1TVNGGGVGVINNGKKK176 pKa = 8.49KK177 pKa = 10.55KKK179 pKa = 10.22RR180 pKa = 11.84DDD182 pKa = 3.94DD183 pKa = 3.81SVFSFVKKK191 pKa = 10.35DDD193 pKa = 5.02IDDD196 pKa = 4.07DDD198 pKa = 3.54SSYYY202 pKa = 10.26QQSIFDDD209 pKa = 4.16ASVTLPTFYYY219 pKa = 11.18YY220 pKa = 9.46TADDD224 pKa = 4.43DD225 pKa = 4.36DD226 pKa = 4.57EEE228 pKa = 4.57DDD230 pKa = 3.62TVPEEE235 pKa = 4.67QTITINSDDD244 pKa = 2.92YYY246 pKa = 11.92DD247 pKa = 3.43DD248 pKa = 4.37KKK250 pKa = 10.75KKK252 pKa = 11.04KK253 pKa = 9.82GTLIIDDD260 pKa = 3.6PEEE263 pKa = 4.0YYY265 pKa = 9.48HHH267 pKa = 7.2KK268 pKa = 10.41DDD270 pKa = 4.59EEE272 pKa = 4.94DD273 pKa = 3.82ATIIFNQSCSVMIDDD288 pKa = 2.96YY289 pKa = 11.52DD290 pKa = 3.48DDD292 pKa = 4.16NDDD295 pKa = 4.05CLIDDD300 pKa = 3.83NGYYY304 pKa = 8.85VVFYYY309 pKa = 10.5EEE311 pKa = 4.5KK312 pKa = 10.43KK313 pKa = 9.65KKK315 pKa = 9.51DDD317 pKa = 3.19GTTIDDD323 pKa = 3.82NIYYY327 pKa = 7.32QKKK330 pKa = 9.29EE331 pKa = 3.89HHH333 pKa = 6.53KKK335 pKa = 10.42YYY337 pKa = 10.41NNHHH341 pKa = 3.92TMMGLFISEEE351 pKa = 4.05YY352 pKa = 9.81KK353 pKa = 10.21KKK355 pKa = 10.6EEE357 pKa = 5.25KK358 pKa = 9.69YY359 pKa = 8.18DDD361 pKa = 3.87YYY363 pKa = 10.39STICEEE369 pKa = 4.39IPAPSNPLPCNDDD382 pKa = 5.56DD383 pKa = 5.41DD384 pKa = 7.38DD385 pKa = 7.62DD386 pKa = 7.72DD387 pKa = 7.58DD388 pKa = 7.65DD389 pKa = 7.65DD390 pKa = 7.65DD391 pKa = 7.64DD392 pKa = 7.49DD393 pKa = 7.59DD394 pKa = 7.3DD395 pKa = 5.79DD396 pKa = 6.17DD397 pKa = 6.2VGQVNTIDDD406 pKa = 4.87YYY408 pKa = 10.35VPAKKK413 pKa = 10.24EEE415 pKa = 4.12NINLVSQTHHH425 pKa = 5.81KKK427 pKa = 9.96KKK429 pKa = 10.55YY430 pKa = 9.42ISNIRR435 pKa = 11.84GTTVISKKK443 pKa = 9.43EE444 pKa = 4.14IVTKKK449 pKa = 10.53NNSLSVDDD457 pKa = 3.5TKKK460 pKa = 10.87PEEE463 pKa = 4.01EEE465 pKa = 4.45YY466 pKa = 10.3FTIVLDDD473 pKa = 4.06NRR475 pKa = 11.84LQKKK479 pKa = 10.92KK480 pKa = 9.54IKKK483 pKa = 10.28SN
DDD2 pKa = 5.84IDDD5 pKa = 3.66NCDDD9 pKa = 3.07NIDDD13 pKa = 3.75EE14 pKa = 6.35FDDD17 pKa = 3.4DDD19 pKa = 3.97DDD21 pKa = 4.02FTACQGDDD29 pKa = 4.7DDD31 pKa = 5.45DD32 pKa = 5.3DDD34 pKa = 4.19NSYYY38 pKa = 10.12GAPEEE43 pKa = 3.83CDDD46 pKa = 5.83IDDD49 pKa = 3.54NCDDD53 pKa = 3.27LVDDD57 pKa = 4.18DDD59 pKa = 4.23DD60 pKa = 3.87SVIGLPDDD68 pKa = 3.3VITEEE73 pKa = 4.18PLGDDD78 pKa = 3.84CQGVAEEE85 pKa = 4.25TVDDD89 pKa = 3.53LNASDDD95 pKa = 4.74SSISYYY101 pKa = 9.43WSNGSTDDD109 pKa = 2.79SIIVSDDD116 pKa = 3.57NTYYY120 pKa = 8.97VSVTGNSGCTTTSSYYY136 pKa = 10.75STAVPSDDD144 pKa = 3.61LSGYYY149 pKa = 10.36MLANKKK155 pKa = 9.68EE156 pKa = 4.13YYY158 pKa = 10.47KKK160 pKa = 9.62EE161 pKa = 4.1TVNGGGVGVINNGKKK176 pKa = 8.49KK177 pKa = 10.55KKK179 pKa = 10.22RR180 pKa = 11.84DDD182 pKa = 3.94DD183 pKa = 3.81SVFSFVKKK191 pKa = 10.35DDD193 pKa = 5.02IDDD196 pKa = 4.07DDD198 pKa = 3.54SSYYY202 pKa = 10.26QQSIFDDD209 pKa = 4.16ASVTLPTFYYY219 pKa = 11.18YY220 pKa = 9.46TADDD224 pKa = 4.43DD225 pKa = 4.36DD226 pKa = 4.57EEE228 pKa = 4.57DDD230 pKa = 3.62TVPEEE235 pKa = 4.67QTITINSDDD244 pKa = 2.92YYY246 pKa = 11.92DD247 pKa = 3.43DD248 pKa = 4.37KKK250 pKa = 10.75KKK252 pKa = 11.04KK253 pKa = 9.82GTLIIDDD260 pKa = 3.6PEEE263 pKa = 4.0YYY265 pKa = 9.48HHH267 pKa = 7.2KK268 pKa = 10.41DDD270 pKa = 4.59EEE272 pKa = 4.94DD273 pKa = 3.82ATIIFNQSCSVMIDDD288 pKa = 2.96YY289 pKa = 11.52DD290 pKa = 3.48DDD292 pKa = 4.16NDDD295 pKa = 4.05CLIDDD300 pKa = 3.83NGYYY304 pKa = 8.85VVFYYY309 pKa = 10.5EEE311 pKa = 4.5KK312 pKa = 10.43KK313 pKa = 9.65KKK315 pKa = 9.51DDD317 pKa = 3.19GTTIDDD323 pKa = 3.82NIYYY327 pKa = 7.32QKKK330 pKa = 9.29EE331 pKa = 3.89HHH333 pKa = 6.53KKK335 pKa = 10.42YYY337 pKa = 10.41NNHHH341 pKa = 3.92TMMGLFISEEE351 pKa = 4.05YY352 pKa = 9.81KK353 pKa = 10.21KKK355 pKa = 10.6EEE357 pKa = 5.25KK358 pKa = 9.69YY359 pKa = 8.18DDD361 pKa = 3.87YYY363 pKa = 10.39STICEEE369 pKa = 4.39IPAPSNPLPCNDDD382 pKa = 5.56DD383 pKa = 5.41DD384 pKa = 7.38DD385 pKa = 7.62DD386 pKa = 7.72DD387 pKa = 7.58DD388 pKa = 7.65DD389 pKa = 7.65DD390 pKa = 7.65DD391 pKa = 7.64DD392 pKa = 7.49DD393 pKa = 7.59DD394 pKa = 7.3DD395 pKa = 5.79DD396 pKa = 6.17DD397 pKa = 6.2VGQVNTIDDD406 pKa = 4.87YYY408 pKa = 10.35VPAKKK413 pKa = 10.24EEE415 pKa = 4.12NINLVSQTHHH425 pKa = 5.81KKK427 pKa = 9.96KKK429 pKa = 10.55YY430 pKa = 9.42ISNIRR435 pKa = 11.84GTTVISKKK443 pKa = 9.43EE444 pKa = 4.14IVTKKK449 pKa = 10.53NNSLSVDDD457 pKa = 3.5TKKK460 pKa = 10.87PEEE463 pKa = 4.01EEE465 pKa = 4.45YY466 pKa = 10.3FTIVLDDD473 pKa = 4.06NRR475 pKa = 11.84LQKKK479 pKa = 10.92KK480 pKa = 9.54IKKK483 pKa = 10.28SN
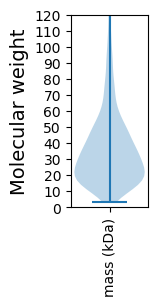
Molecular weight: 53.04 kDa
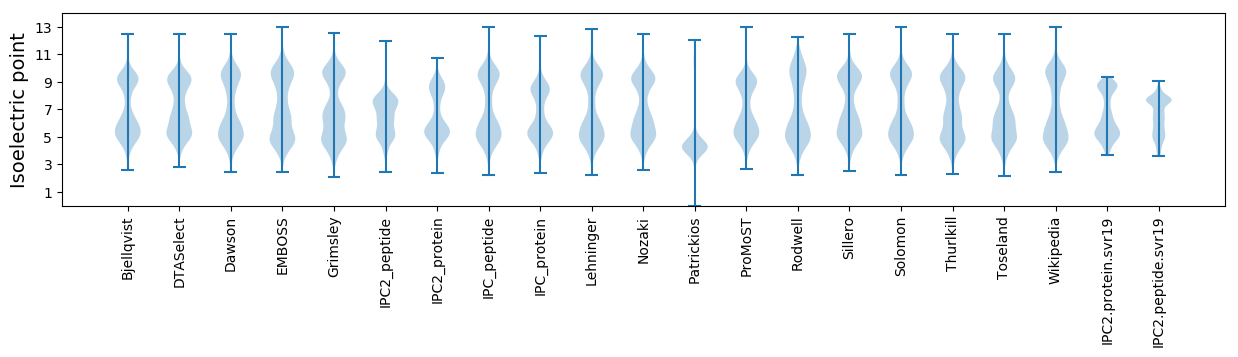
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A327RL91|A0A327RL91_9FLAO Glycine/D-amino acid oxidase-like deaminating enzyme OS=Olleya aquimaris OX=639310 GN=LY08_00048 PE=4 SV=1
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
MM1 pKa = 8.0PKK3 pKa = 9.06RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.0KK42 pKa = 10.66LSVSTEE48 pKa = 3.92TRR50 pKa = 11.84HH51 pKa = 6.44KK52 pKa = 10.57KK53 pKa = 9.81
Molecular weight: 6.35 kDa
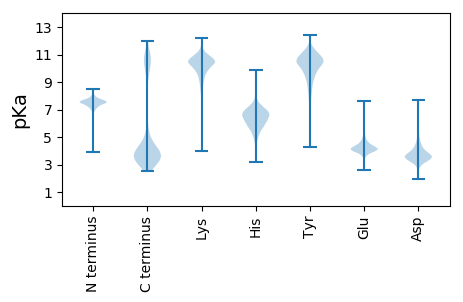
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
899899 |
27 |
2399 |
340.6 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.296 ± 0.048 | 0.752 ± 0.015 |
5.955 ± 0.037 | 6.222 ± 0.05 |
5.229 ± 0.039 | 6.16 ± 0.048 |
1.734 ± 0.021 | 7.927 ± 0.042 |
7.782 ± 0.063 | 9.257 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.007 ± 0.023 | 6.691 ± 0.055 |
3.296 ± 0.029 | 3.663 ± 0.029 |
3.076 ± 0.029 | 6.294 ± 0.04 |
6.339 ± 0.05 | 6.278 ± 0.035 |
0.97 ± 0.015 | 4.072 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
