
Cohaesibacter sp. CAU 1516
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Cohaesibacteraceae; Cohaesibacter; unclassified Cohaesibacter
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
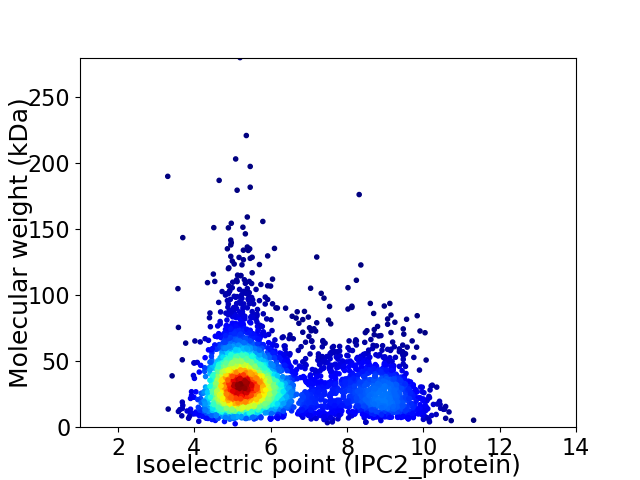
Virtual 2D-PAGE plot for 3998 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
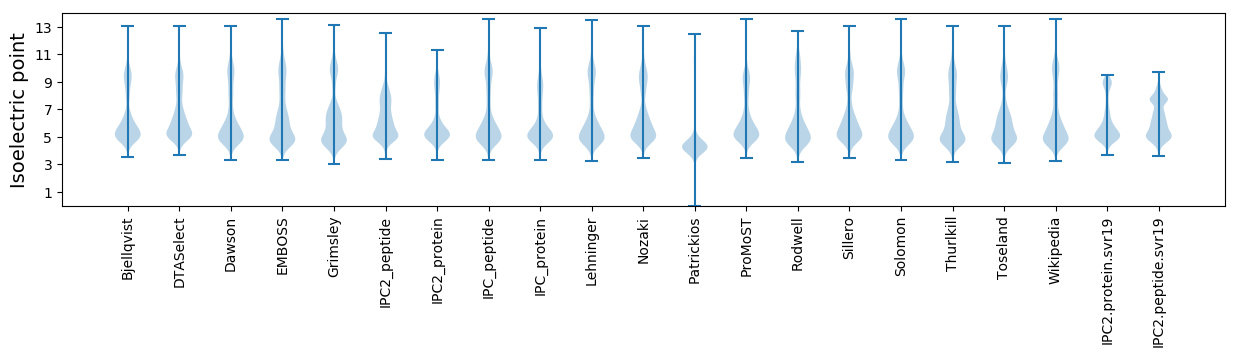
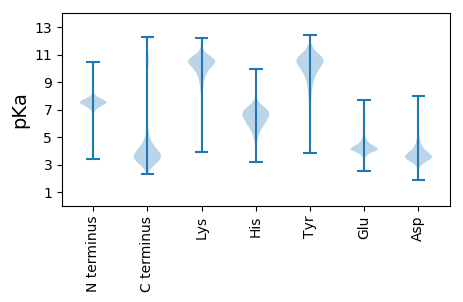
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5R8YG09|A0A5R8YG09_9RHIZ LysR family transcriptional regulator OS=Cohaesibacter sp. CAU 1516 OX=2576038 GN=FDK21_06130 PE=3 SV=1
MM1 pKa = 7.38CTLCGIGLPDD11 pKa = 3.36FHH13 pKa = 6.98GQTFTPLSGSEE24 pKa = 4.11SGDD27 pKa = 3.24GLTASTASVSDD38 pKa = 4.04PGEE41 pKa = 3.78VWKK44 pKa = 10.93GVLHH48 pKa = 5.34DD49 pKa = 4.23TKK51 pKa = 10.32WAVSSLQFSFPTSASFYY68 pKa = 9.1ASTYY72 pKa = 10.57GSEE75 pKa = 4.16QPDD78 pKa = 3.64LGFEE82 pKa = 4.38ALNSVQKK89 pKa = 10.81DD90 pKa = 3.7AVRR93 pKa = 11.84SLYY96 pKa = 10.94NEE98 pKa = 3.93IEE100 pKa = 4.35SFTDD104 pKa = 3.34LTFTEE109 pKa = 4.69VTEE112 pKa = 4.46SSTTHH117 pKa = 6.11AEE119 pKa = 3.69LRR121 pKa = 11.84FAMTLNNTPTAAGYY135 pKa = 9.45FPYY138 pKa = 10.74GSDD141 pKa = 3.32EE142 pKa = 5.84AGDD145 pKa = 3.04SWYY148 pKa = 10.9RR149 pKa = 11.84NDD151 pKa = 5.02GNYY154 pKa = 10.4DD155 pKa = 3.5DD156 pKa = 5.42PRR158 pKa = 11.84LGSYY162 pKa = 10.22AYY164 pKa = 10.61NEE166 pKa = 4.26GFMHH170 pKa = 7.38EE171 pKa = 4.27IGHH174 pKa = 5.52TLGLTHH180 pKa = 6.53GHH182 pKa = 7.27DD183 pKa = 3.53SGNPFGALPSNVDD196 pKa = 3.55SNEE199 pKa = 3.78FSVMTYY205 pKa = 9.61RR206 pKa = 11.84DD207 pKa = 3.54HH208 pKa = 9.04VGDD211 pKa = 4.07SFTDD215 pKa = 3.84PSEE218 pKa = 4.22NEE220 pKa = 3.7IGGNAQSYY228 pKa = 8.41MMYY231 pKa = 10.38DD232 pKa = 3.56IAALQYY238 pKa = 9.99MYY240 pKa = 10.93GADD243 pKa = 3.61YY244 pKa = 10.71SASGNVWSGDD254 pKa = 3.23TVYY257 pKa = 10.79TFDD260 pKa = 4.44PSSGQMSINGAGQGTPVEE278 pKa = 3.97NRR280 pKa = 11.84IFRR283 pKa = 11.84TIWDD287 pKa = 3.81GNGVDD292 pKa = 4.87TYY294 pKa = 11.69DD295 pKa = 3.5LSNYY299 pKa = 7.11TTDD302 pKa = 5.02LSINLNPGEE311 pKa = 3.99WSIFSQAQLADD322 pKa = 3.99LNGGSRR328 pKa = 11.84SDD330 pKa = 3.42YY331 pKa = 10.33LARR334 pKa = 11.84GNVANALLFEE344 pKa = 5.07GDD346 pKa = 3.68LRR348 pKa = 11.84SLIEE352 pKa = 3.89NANGGRR358 pKa = 11.84GNDD361 pKa = 3.89TIIGNQGSNEE371 pKa = 3.63LRR373 pKa = 11.84GNGGNDD379 pKa = 3.02NLQGGDD385 pKa = 3.58GRR387 pKa = 11.84DD388 pKa = 3.11KK389 pKa = 11.05AIYY392 pKa = 9.35SGAYY396 pKa = 8.81SEE398 pKa = 4.75YY399 pKa = 10.72SITEE403 pKa = 4.33NIDD406 pKa = 3.05GSFTIAHH413 pKa = 6.24TGGDD417 pKa = 3.66RR418 pKa = 11.84SDD420 pKa = 3.64GTDD423 pKa = 2.88TVRR426 pKa = 11.84DD427 pKa = 3.03IEE429 pKa = 4.37YY430 pKa = 10.61AVFSDD435 pKa = 4.76RR436 pKa = 11.84EE437 pKa = 4.06VALAPLTGPLDD448 pKa = 3.76VVFLQDD454 pKa = 3.68LTASFIDD461 pKa = 4.56DD462 pKa = 4.3LPSMRR467 pKa = 11.84AAVDD471 pKa = 4.4DD472 pKa = 3.94IAAVIQNAYY481 pKa = 9.75ANSRR485 pKa = 11.84FAVTSFRR492 pKa = 11.84DD493 pKa = 3.29TGDD496 pKa = 3.25SYY498 pKa = 11.57VYY500 pKa = 10.16RR501 pKa = 11.84VEE503 pKa = 5.22ADD505 pKa = 3.68FTQSTSSLVSTYY517 pKa = 11.25NSMNATGGGDD527 pKa = 3.36TPEE530 pKa = 4.32AQLTALLQASTDD542 pKa = 3.32SSLSYY547 pKa = 10.7QSGSTRR553 pKa = 11.84IFILATDD560 pKa = 4.2AGYY563 pKa = 10.87HH564 pKa = 5.52NYY566 pKa = 10.38EE567 pKa = 4.68SIASIAALMASNDD580 pKa = 3.99IIPIFAVASGEE591 pKa = 4.03VSTYY595 pKa = 10.89QNLANEE601 pKa = 4.92LGTGVVVTIEE611 pKa = 3.99SDD613 pKa = 3.51SADD616 pKa = 3.42FADD619 pKa = 4.41SIRR622 pKa = 11.84YY623 pKa = 8.74ALASIDD629 pKa = 4.27AEE631 pKa = 4.29VTEE634 pKa = 5.34AGTSSGDD641 pKa = 3.39TLAGGDD647 pKa = 3.98TADD650 pKa = 4.69GIYY653 pKa = 10.89GLGGDD658 pKa = 4.55DD659 pKa = 3.95TISGNGGDD667 pKa = 4.76DD668 pKa = 3.98LLDD671 pKa = 4.19GGSGNDD677 pKa = 3.55SLLGGDD683 pKa = 5.79DD684 pKa = 3.77NDD686 pKa = 3.7QLLGGSGNDD695 pKa = 3.19NLLGGEE701 pKa = 4.31GNDD704 pKa = 3.41RR705 pKa = 11.84LVGGDD710 pKa = 3.79GDD712 pKa = 5.31DD713 pKa = 3.72VLLGDD718 pKa = 4.83GDD720 pKa = 4.19SSSSLFPSSLSLNSLFTLGFDD741 pKa = 4.28EE742 pKa = 6.54DD743 pKa = 4.02IQNSTTQAHH752 pKa = 5.82ATVQGTGSGSVNYY765 pKa = 10.4YY766 pKa = 10.38SFTVAAAGLSVVLDD780 pKa = 3.6IDD782 pKa = 4.36YY783 pKa = 11.27ADD785 pKa = 3.79QNIGASDD792 pKa = 3.87FDD794 pKa = 3.81SWIVLYY800 pKa = 10.66DD801 pKa = 3.35SSGNQLSYY809 pKa = 11.55SDD811 pKa = 5.12DD812 pKa = 4.35DD813 pKa = 4.36YY814 pKa = 12.03GGDD817 pKa = 3.58PGSVDD822 pKa = 2.98SHH824 pKa = 7.41DD825 pKa = 4.53SYY827 pKa = 11.7LSHH830 pKa = 6.96TLGSAGTYY838 pKa = 8.34YY839 pKa = 10.81VAVGSYY845 pKa = 10.72SSLSPIPVGGTYY857 pKa = 9.85EE858 pKa = 3.96LHH860 pKa = 6.29VSVNGAIGEE869 pKa = 4.29PPTVGAGNDD878 pKa = 3.52VLNGGEE884 pKa = 4.8GADD887 pKa = 3.91SMAGGAGNDD896 pKa = 3.23VYY898 pKa = 11.62YY899 pKa = 10.71VDD901 pKa = 4.66NVGDD905 pKa = 4.13TIEE908 pKa = 4.31EE909 pKa = 4.1NADD912 pKa = 3.39NGNDD916 pKa = 3.5LVFSYY921 pKa = 10.8ISLALRR927 pKa = 11.84DD928 pKa = 3.65HH929 pKa = 6.07SQNLEE934 pKa = 3.99HH935 pKa = 7.29LSLLGPGNINGTGNGLGNTITGNSGNNVLNGAYY968 pKa = 10.32GNDD971 pKa = 3.54VLRR974 pKa = 11.84GGAGNDD980 pKa = 2.97IFKK983 pKa = 10.82DD984 pKa = 3.59DD985 pKa = 3.65VGADD989 pKa = 3.5RR990 pKa = 11.84MIGGSGNDD998 pKa = 3.14VYY1000 pKa = 11.67YY1001 pKa = 10.74VDD1003 pKa = 4.18NVGDD1007 pKa = 3.81RR1008 pKa = 11.84VEE1010 pKa = 4.31EE1011 pKa = 4.29SAGSGTDD1018 pKa = 3.1QVFSYY1023 pKa = 10.89ISFALRR1029 pKa = 11.84DD1030 pKa = 3.43HH1031 pKa = 5.98SQYY1034 pKa = 11.19LEE1036 pKa = 4.02HH1037 pKa = 7.28LSLLGSGNTNGTGNAQHH1054 pKa = 5.94NTITGNSGNNILNGAGGNDD1073 pKa = 3.45ILRR1076 pKa = 11.84GGAGNDD1082 pKa = 2.91IFKK1085 pKa = 11.1DD1086 pKa = 3.56NIGADD1091 pKa = 3.49RR1092 pKa = 11.84MIGGTGNDD1100 pKa = 3.08VYY1102 pKa = 11.54YY1103 pKa = 10.84VDD1105 pKa = 4.03NGGDD1109 pKa = 3.35RR1110 pKa = 11.84VEE1112 pKa = 4.5EE1113 pKa = 4.28SAGSGTDD1120 pKa = 3.1QVFSYY1125 pKa = 10.89ISFALRR1131 pKa = 11.84DD1132 pKa = 3.43HH1133 pKa = 5.98SQYY1136 pKa = 11.19LEE1138 pKa = 4.02HH1139 pKa = 7.28LSLLGSGDD1147 pKa = 3.36INGTGNGLGNTITGNAGNNVLNGAYY1172 pKa = 10.33GNDD1175 pKa = 3.89LLRR1178 pKa = 11.84GGAGNDD1184 pKa = 2.99IFKK1187 pKa = 10.6DD1188 pKa = 3.6DD1189 pKa = 3.9NGADD1193 pKa = 3.51RR1194 pKa = 11.84MIGGTGNDD1202 pKa = 3.14VYY1204 pKa = 11.59YY1205 pKa = 10.67VDD1207 pKa = 4.18NVGDD1211 pKa = 3.81RR1212 pKa = 11.84VEE1214 pKa = 4.31EE1215 pKa = 4.29SAGSGTDD1222 pKa = 3.11QVFSYY1227 pKa = 11.57VSFTLRR1233 pKa = 11.84NHH1235 pKa = 5.25SQYY1238 pKa = 11.15LEE1240 pKa = 4.06HH1241 pKa = 7.32LSLLGAGNINGTGNGLNNIITGNSGNNVLNGAYY1274 pKa = 10.05GQDD1277 pKa = 3.29ILRR1280 pKa = 11.84GGAGNDD1286 pKa = 2.87IFFDD1290 pKa = 4.39DD1291 pKa = 4.24SGNDD1295 pKa = 2.82IFTGGTGADD1304 pKa = 3.11TFIFQRR1310 pKa = 11.84SGEE1313 pKa = 3.99FDD1315 pKa = 3.81IITDD1319 pKa = 4.24FEE1321 pKa = 5.29DD1322 pKa = 5.41GIDD1325 pKa = 3.86TIRR1328 pKa = 11.84IATGSNTFSQVKK1340 pKa = 10.12ISDD1343 pKa = 4.21DD1344 pKa = 3.71GADD1347 pKa = 3.76TIVQFDD1353 pKa = 3.88GSTVVLNNFNHH1364 pKa = 6.4TLLGADD1370 pKa = 4.08DD1371 pKa = 4.45FSFVV1375 pKa = 3.42
MM1 pKa = 7.38CTLCGIGLPDD11 pKa = 3.36FHH13 pKa = 6.98GQTFTPLSGSEE24 pKa = 4.11SGDD27 pKa = 3.24GLTASTASVSDD38 pKa = 4.04PGEE41 pKa = 3.78VWKK44 pKa = 10.93GVLHH48 pKa = 5.34DD49 pKa = 4.23TKK51 pKa = 10.32WAVSSLQFSFPTSASFYY68 pKa = 9.1ASTYY72 pKa = 10.57GSEE75 pKa = 4.16QPDD78 pKa = 3.64LGFEE82 pKa = 4.38ALNSVQKK89 pKa = 10.81DD90 pKa = 3.7AVRR93 pKa = 11.84SLYY96 pKa = 10.94NEE98 pKa = 3.93IEE100 pKa = 4.35SFTDD104 pKa = 3.34LTFTEE109 pKa = 4.69VTEE112 pKa = 4.46SSTTHH117 pKa = 6.11AEE119 pKa = 3.69LRR121 pKa = 11.84FAMTLNNTPTAAGYY135 pKa = 9.45FPYY138 pKa = 10.74GSDD141 pKa = 3.32EE142 pKa = 5.84AGDD145 pKa = 3.04SWYY148 pKa = 10.9RR149 pKa = 11.84NDD151 pKa = 5.02GNYY154 pKa = 10.4DD155 pKa = 3.5DD156 pKa = 5.42PRR158 pKa = 11.84LGSYY162 pKa = 10.22AYY164 pKa = 10.61NEE166 pKa = 4.26GFMHH170 pKa = 7.38EE171 pKa = 4.27IGHH174 pKa = 5.52TLGLTHH180 pKa = 6.53GHH182 pKa = 7.27DD183 pKa = 3.53SGNPFGALPSNVDD196 pKa = 3.55SNEE199 pKa = 3.78FSVMTYY205 pKa = 9.61RR206 pKa = 11.84DD207 pKa = 3.54HH208 pKa = 9.04VGDD211 pKa = 4.07SFTDD215 pKa = 3.84PSEE218 pKa = 4.22NEE220 pKa = 3.7IGGNAQSYY228 pKa = 8.41MMYY231 pKa = 10.38DD232 pKa = 3.56IAALQYY238 pKa = 9.99MYY240 pKa = 10.93GADD243 pKa = 3.61YY244 pKa = 10.71SASGNVWSGDD254 pKa = 3.23TVYY257 pKa = 10.79TFDD260 pKa = 4.44PSSGQMSINGAGQGTPVEE278 pKa = 3.97NRR280 pKa = 11.84IFRR283 pKa = 11.84TIWDD287 pKa = 3.81GNGVDD292 pKa = 4.87TYY294 pKa = 11.69DD295 pKa = 3.5LSNYY299 pKa = 7.11TTDD302 pKa = 5.02LSINLNPGEE311 pKa = 3.99WSIFSQAQLADD322 pKa = 3.99LNGGSRR328 pKa = 11.84SDD330 pKa = 3.42YY331 pKa = 10.33LARR334 pKa = 11.84GNVANALLFEE344 pKa = 5.07GDD346 pKa = 3.68LRR348 pKa = 11.84SLIEE352 pKa = 3.89NANGGRR358 pKa = 11.84GNDD361 pKa = 3.89TIIGNQGSNEE371 pKa = 3.63LRR373 pKa = 11.84GNGGNDD379 pKa = 3.02NLQGGDD385 pKa = 3.58GRR387 pKa = 11.84DD388 pKa = 3.11KK389 pKa = 11.05AIYY392 pKa = 9.35SGAYY396 pKa = 8.81SEE398 pKa = 4.75YY399 pKa = 10.72SITEE403 pKa = 4.33NIDD406 pKa = 3.05GSFTIAHH413 pKa = 6.24TGGDD417 pKa = 3.66RR418 pKa = 11.84SDD420 pKa = 3.64GTDD423 pKa = 2.88TVRR426 pKa = 11.84DD427 pKa = 3.03IEE429 pKa = 4.37YY430 pKa = 10.61AVFSDD435 pKa = 4.76RR436 pKa = 11.84EE437 pKa = 4.06VALAPLTGPLDD448 pKa = 3.76VVFLQDD454 pKa = 3.68LTASFIDD461 pKa = 4.56DD462 pKa = 4.3LPSMRR467 pKa = 11.84AAVDD471 pKa = 4.4DD472 pKa = 3.94IAAVIQNAYY481 pKa = 9.75ANSRR485 pKa = 11.84FAVTSFRR492 pKa = 11.84DD493 pKa = 3.29TGDD496 pKa = 3.25SYY498 pKa = 11.57VYY500 pKa = 10.16RR501 pKa = 11.84VEE503 pKa = 5.22ADD505 pKa = 3.68FTQSTSSLVSTYY517 pKa = 11.25NSMNATGGGDD527 pKa = 3.36TPEE530 pKa = 4.32AQLTALLQASTDD542 pKa = 3.32SSLSYY547 pKa = 10.7QSGSTRR553 pKa = 11.84IFILATDD560 pKa = 4.2AGYY563 pKa = 10.87HH564 pKa = 5.52NYY566 pKa = 10.38EE567 pKa = 4.68SIASIAALMASNDD580 pKa = 3.99IIPIFAVASGEE591 pKa = 4.03VSTYY595 pKa = 10.89QNLANEE601 pKa = 4.92LGTGVVVTIEE611 pKa = 3.99SDD613 pKa = 3.51SADD616 pKa = 3.42FADD619 pKa = 4.41SIRR622 pKa = 11.84YY623 pKa = 8.74ALASIDD629 pKa = 4.27AEE631 pKa = 4.29VTEE634 pKa = 5.34AGTSSGDD641 pKa = 3.39TLAGGDD647 pKa = 3.98TADD650 pKa = 4.69GIYY653 pKa = 10.89GLGGDD658 pKa = 4.55DD659 pKa = 3.95TISGNGGDD667 pKa = 4.76DD668 pKa = 3.98LLDD671 pKa = 4.19GGSGNDD677 pKa = 3.55SLLGGDD683 pKa = 5.79DD684 pKa = 3.77NDD686 pKa = 3.7QLLGGSGNDD695 pKa = 3.19NLLGGEE701 pKa = 4.31GNDD704 pKa = 3.41RR705 pKa = 11.84LVGGDD710 pKa = 3.79GDD712 pKa = 5.31DD713 pKa = 3.72VLLGDD718 pKa = 4.83GDD720 pKa = 4.19SSSSLFPSSLSLNSLFTLGFDD741 pKa = 4.28EE742 pKa = 6.54DD743 pKa = 4.02IQNSTTQAHH752 pKa = 5.82ATVQGTGSGSVNYY765 pKa = 10.4YY766 pKa = 10.38SFTVAAAGLSVVLDD780 pKa = 3.6IDD782 pKa = 4.36YY783 pKa = 11.27ADD785 pKa = 3.79QNIGASDD792 pKa = 3.87FDD794 pKa = 3.81SWIVLYY800 pKa = 10.66DD801 pKa = 3.35SSGNQLSYY809 pKa = 11.55SDD811 pKa = 5.12DD812 pKa = 4.35DD813 pKa = 4.36YY814 pKa = 12.03GGDD817 pKa = 3.58PGSVDD822 pKa = 2.98SHH824 pKa = 7.41DD825 pKa = 4.53SYY827 pKa = 11.7LSHH830 pKa = 6.96TLGSAGTYY838 pKa = 8.34YY839 pKa = 10.81VAVGSYY845 pKa = 10.72SSLSPIPVGGTYY857 pKa = 9.85EE858 pKa = 3.96LHH860 pKa = 6.29VSVNGAIGEE869 pKa = 4.29PPTVGAGNDD878 pKa = 3.52VLNGGEE884 pKa = 4.8GADD887 pKa = 3.91SMAGGAGNDD896 pKa = 3.23VYY898 pKa = 11.62YY899 pKa = 10.71VDD901 pKa = 4.66NVGDD905 pKa = 4.13TIEE908 pKa = 4.31EE909 pKa = 4.1NADD912 pKa = 3.39NGNDD916 pKa = 3.5LVFSYY921 pKa = 10.8ISLALRR927 pKa = 11.84DD928 pKa = 3.65HH929 pKa = 6.07SQNLEE934 pKa = 3.99HH935 pKa = 7.29LSLLGPGNINGTGNGLGNTITGNSGNNVLNGAYY968 pKa = 10.32GNDD971 pKa = 3.54VLRR974 pKa = 11.84GGAGNDD980 pKa = 2.97IFKK983 pKa = 10.82DD984 pKa = 3.59DD985 pKa = 3.65VGADD989 pKa = 3.5RR990 pKa = 11.84MIGGSGNDD998 pKa = 3.14VYY1000 pKa = 11.67YY1001 pKa = 10.74VDD1003 pKa = 4.18NVGDD1007 pKa = 3.81RR1008 pKa = 11.84VEE1010 pKa = 4.31EE1011 pKa = 4.29SAGSGTDD1018 pKa = 3.1QVFSYY1023 pKa = 10.89ISFALRR1029 pKa = 11.84DD1030 pKa = 3.43HH1031 pKa = 5.98SQYY1034 pKa = 11.19LEE1036 pKa = 4.02HH1037 pKa = 7.28LSLLGSGNTNGTGNAQHH1054 pKa = 5.94NTITGNSGNNILNGAGGNDD1073 pKa = 3.45ILRR1076 pKa = 11.84GGAGNDD1082 pKa = 2.91IFKK1085 pKa = 11.1DD1086 pKa = 3.56NIGADD1091 pKa = 3.49RR1092 pKa = 11.84MIGGTGNDD1100 pKa = 3.08VYY1102 pKa = 11.54YY1103 pKa = 10.84VDD1105 pKa = 4.03NGGDD1109 pKa = 3.35RR1110 pKa = 11.84VEE1112 pKa = 4.5EE1113 pKa = 4.28SAGSGTDD1120 pKa = 3.1QVFSYY1125 pKa = 10.89ISFALRR1131 pKa = 11.84DD1132 pKa = 3.43HH1133 pKa = 5.98SQYY1136 pKa = 11.19LEE1138 pKa = 4.02HH1139 pKa = 7.28LSLLGSGDD1147 pKa = 3.36INGTGNGLGNTITGNAGNNVLNGAYY1172 pKa = 10.33GNDD1175 pKa = 3.89LLRR1178 pKa = 11.84GGAGNDD1184 pKa = 2.99IFKK1187 pKa = 10.6DD1188 pKa = 3.6DD1189 pKa = 3.9NGADD1193 pKa = 3.51RR1194 pKa = 11.84MIGGTGNDD1202 pKa = 3.14VYY1204 pKa = 11.59YY1205 pKa = 10.67VDD1207 pKa = 4.18NVGDD1211 pKa = 3.81RR1212 pKa = 11.84VEE1214 pKa = 4.31EE1215 pKa = 4.29SAGSGTDD1222 pKa = 3.11QVFSYY1227 pKa = 11.57VSFTLRR1233 pKa = 11.84NHH1235 pKa = 5.25SQYY1238 pKa = 11.15LEE1240 pKa = 4.06HH1241 pKa = 7.32LSLLGAGNINGTGNGLNNIITGNSGNNVLNGAYY1274 pKa = 10.05GQDD1277 pKa = 3.29ILRR1280 pKa = 11.84GGAGNDD1286 pKa = 2.87IFFDD1290 pKa = 4.39DD1291 pKa = 4.24SGNDD1295 pKa = 2.82IFTGGTGADD1304 pKa = 3.11TFIFQRR1310 pKa = 11.84SGEE1313 pKa = 3.99FDD1315 pKa = 3.81IITDD1319 pKa = 4.24FEE1321 pKa = 5.29DD1322 pKa = 5.41GIDD1325 pKa = 3.86TIRR1328 pKa = 11.84IATGSNTFSQVKK1340 pKa = 10.12ISDD1343 pKa = 4.21DD1344 pKa = 3.71GADD1347 pKa = 3.76TIVQFDD1353 pKa = 3.88GSTVVLNNFNHH1364 pKa = 6.4TLLGADD1370 pKa = 4.08DD1371 pKa = 4.45FSFVV1375 pKa = 3.42
Molecular weight: 143.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5R8YD88|A0A5R8YD88_9RHIZ Uncharacterized protein OS=Cohaesibacter sp. CAU 1516 OX=2576038 GN=FDK21_00855 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.48GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1310659 |
24 |
2583 |
327.8 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.855 ± 0.044 | 0.966 ± 0.012 |
6.099 ± 0.037 | 5.916 ± 0.038 |
3.947 ± 0.025 | 7.961 ± 0.038 |
2.17 ± 0.02 | 5.9 ± 0.029 |
4.37 ± 0.031 | 10.247 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.872 ± 0.02 | 3.063 ± 0.023 |
4.602 ± 0.03 | 3.528 ± 0.025 |
5.806 ± 0.037 | 6.017 ± 0.033 |
5.164 ± 0.03 | 6.911 ± 0.032 |
1.245 ± 0.014 | 2.36 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
