
Gonium pectorale (Green alga)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; core chlorophytes; Chlorophyceae; CS clade; Chlamydomonadales; Volvocaceae; Gonium
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
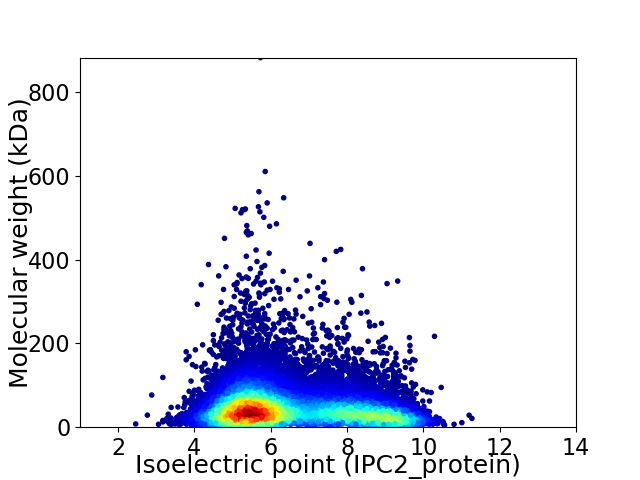
Virtual 2D-PAGE plot for 16224 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A150H2N5|A0A150H2N5_GONPE Prolyl aminopeptidase OS=Gonium pectorale OX=33097 GN=GPECTOR_2g980 PE=4 SV=1
MM1 pKa = 6.94NTIEE5 pKa = 5.37DD6 pKa = 4.12DD7 pKa = 4.23CAHH10 pKa = 6.74HH11 pKa = 6.95ALLPEE16 pKa = 4.33GHH18 pKa = 5.91STLYY22 pKa = 10.74DD23 pKa = 3.69ALPSFDD29 pKa = 4.61EE30 pKa = 4.9DD31 pKa = 3.48LSEE34 pKa = 4.74DD35 pKa = 3.92VIVPLSTPPEE45 pKa = 4.16TEE47 pKa = 4.11PVDD50 pKa = 3.89EE51 pKa = 4.39THH53 pKa = 6.87GGASSSLPATPAEE66 pKa = 4.54RR67 pKa = 11.84PCTFCPASLPLPPASGPSRR86 pKa = 11.84VTLVLDD92 pKa = 4.75LDD94 pKa = 3.93GTLIASEE101 pKa = 4.46EE102 pKa = 3.97LNAANSAFLWNPHH115 pKa = 6.02CGARR119 pKa = 11.84DD120 pKa = 3.56PDD122 pKa = 3.94YY123 pKa = 10.69EE124 pKa = 4.68AMGRR128 pKa = 11.84RR129 pKa = 11.84VWLRR133 pKa = 11.84PGVRR137 pKa = 11.84DD138 pKa = 3.57FLAAVRR144 pKa = 11.84PHH146 pKa = 6.39FEE148 pKa = 3.74IVLFTAATQNWAAAAIEE165 pKa = 3.91QLDD168 pKa = 3.7PSAYY172 pKa = 10.42LFDD175 pKa = 4.07VMLHH179 pKa = 6.58RR180 pKa = 11.84DD181 pKa = 3.46HH182 pKa = 6.48TTSDD186 pKa = 3.71LMWDD190 pKa = 3.71YY191 pKa = 11.88VKK193 pKa = 10.88DD194 pKa = 3.95LSRR197 pKa = 11.84LGRR200 pKa = 11.84DD201 pKa = 3.24LARR204 pKa = 11.84VVIVDD209 pKa = 4.46DD210 pKa = 4.08NPLMFMYY217 pKa = 10.53QPDD220 pKa = 3.79NALHH224 pKa = 6.07IGAYY228 pKa = 8.4EE229 pKa = 4.0AASAGGPDD237 pKa = 3.41NVLEE241 pKa = 4.27RR242 pKa = 11.84VAEE245 pKa = 4.07LLVSKK250 pKa = 10.31VAVAEE255 pKa = 4.15DD256 pKa = 3.48VRR258 pKa = 11.84EE259 pKa = 4.04VLGPLARR266 pKa = 11.84AKK268 pKa = 10.6SCITSTVNAKK278 pKa = 9.7AAPGTAAEE286 pKa = 4.55PLAATASCPAADD298 pKa = 4.29GADD301 pKa = 3.5SAVAAGAAPGDD312 pKa = 3.96GSASTEE318 pKa = 4.0AEE320 pKa = 3.79FDD322 pKa = 3.69GSGAAADD329 pKa = 4.29TDD331 pKa = 4.35TSCDD335 pKa = 3.31AADD338 pKa = 4.15ASNSGAEE345 pKa = 4.21CHH347 pKa = 5.88GAAPEE352 pKa = 4.14GVEE355 pKa = 4.43GEE357 pKa = 4.62EE358 pKa = 4.44EE359 pKa = 4.29VQQDD363 pKa = 4.03EE364 pKa = 4.58QEE366 pKa = 4.31HH367 pKa = 6.14CAGAGDD373 pKa = 4.28EE374 pKa = 4.2EE375 pKa = 5.01LEE377 pKa = 4.46AVDD380 pKa = 5.35SEE382 pKa = 4.58DD383 pKa = 3.55AAHH386 pKa = 6.17VKK388 pKa = 10.25HH389 pKa = 6.93DD390 pKa = 4.3EE391 pKa = 4.15EE392 pKa = 4.33EE393 pKa = 4.22ALPGGDD399 pKa = 3.92LRR401 pKa = 11.84CTEE404 pKa = 4.21GADD407 pKa = 5.0AAFDD411 pKa = 3.64WAGEE415 pKa = 4.09SDD417 pKa = 3.78VPEE420 pKa = 4.58IFDD423 pKa = 3.39EE424 pKa = 4.83HH425 pKa = 6.48GAAACLGADD434 pKa = 4.82SSWQQQSCGGYY445 pKa = 10.56AVDD448 pKa = 5.15APAGEE453 pKa = 4.87DD454 pKa = 3.73SQDD457 pKa = 3.44SAAEE461 pKa = 3.99EE462 pKa = 4.09QPQQHH467 pKa = 6.29NSTTEE472 pKa = 3.81SDD474 pKa = 3.29GHH476 pKa = 7.47DD477 pKa = 4.18DD478 pKa = 5.15FDD480 pKa = 6.55ALMDD484 pKa = 4.44DD485 pKa = 5.75AIAEE489 pKa = 4.31YY490 pKa = 10.77DD491 pKa = 3.21SDD493 pKa = 5.35AYY495 pKa = 10.72DD496 pKa = 3.32GQYY499 pKa = 11.3DD500 pKa = 3.44VDD502 pKa = 3.5IPYY505 pKa = 10.45DD506 pKa = 3.76AEE508 pKa = 4.53CPAGTAAAVASDD520 pKa = 3.46GGAANRR526 pKa = 11.84ASDD529 pKa = 4.61DD530 pKa = 3.86DD531 pKa = 3.9VAEE534 pKa = 5.26AGLQDD539 pKa = 4.9PLRR542 pKa = 11.84QPPVGMVLLADD553 pKa = 4.04LHH555 pKa = 6.62PCLGSNTSGVSPTALLGSKK574 pKa = 9.96RR575 pKa = 11.84ARR577 pKa = 11.84DD578 pKa = 3.47EE579 pKa = 4.81DD580 pKa = 4.28GEE582 pKa = 4.42EE583 pKa = 4.27ACNAAAPHH591 pKa = 6.12CRR593 pKa = 11.84MRR595 pKa = 11.84PAGFACSALEE605 pKa = 4.04TVQVGSCEE613 pKa = 3.93LL614 pKa = 3.47
MM1 pKa = 6.94NTIEE5 pKa = 5.37DD6 pKa = 4.12DD7 pKa = 4.23CAHH10 pKa = 6.74HH11 pKa = 6.95ALLPEE16 pKa = 4.33GHH18 pKa = 5.91STLYY22 pKa = 10.74DD23 pKa = 3.69ALPSFDD29 pKa = 4.61EE30 pKa = 4.9DD31 pKa = 3.48LSEE34 pKa = 4.74DD35 pKa = 3.92VIVPLSTPPEE45 pKa = 4.16TEE47 pKa = 4.11PVDD50 pKa = 3.89EE51 pKa = 4.39THH53 pKa = 6.87GGASSSLPATPAEE66 pKa = 4.54RR67 pKa = 11.84PCTFCPASLPLPPASGPSRR86 pKa = 11.84VTLVLDD92 pKa = 4.75LDD94 pKa = 3.93GTLIASEE101 pKa = 4.46EE102 pKa = 3.97LNAANSAFLWNPHH115 pKa = 6.02CGARR119 pKa = 11.84DD120 pKa = 3.56PDD122 pKa = 3.94YY123 pKa = 10.69EE124 pKa = 4.68AMGRR128 pKa = 11.84RR129 pKa = 11.84VWLRR133 pKa = 11.84PGVRR137 pKa = 11.84DD138 pKa = 3.57FLAAVRR144 pKa = 11.84PHH146 pKa = 6.39FEE148 pKa = 3.74IVLFTAATQNWAAAAIEE165 pKa = 3.91QLDD168 pKa = 3.7PSAYY172 pKa = 10.42LFDD175 pKa = 4.07VMLHH179 pKa = 6.58RR180 pKa = 11.84DD181 pKa = 3.46HH182 pKa = 6.48TTSDD186 pKa = 3.71LMWDD190 pKa = 3.71YY191 pKa = 11.88VKK193 pKa = 10.88DD194 pKa = 3.95LSRR197 pKa = 11.84LGRR200 pKa = 11.84DD201 pKa = 3.24LARR204 pKa = 11.84VVIVDD209 pKa = 4.46DD210 pKa = 4.08NPLMFMYY217 pKa = 10.53QPDD220 pKa = 3.79NALHH224 pKa = 6.07IGAYY228 pKa = 8.4EE229 pKa = 4.0AASAGGPDD237 pKa = 3.41NVLEE241 pKa = 4.27RR242 pKa = 11.84VAEE245 pKa = 4.07LLVSKK250 pKa = 10.31VAVAEE255 pKa = 4.15DD256 pKa = 3.48VRR258 pKa = 11.84EE259 pKa = 4.04VLGPLARR266 pKa = 11.84AKK268 pKa = 10.6SCITSTVNAKK278 pKa = 9.7AAPGTAAEE286 pKa = 4.55PLAATASCPAADD298 pKa = 4.29GADD301 pKa = 3.5SAVAAGAAPGDD312 pKa = 3.96GSASTEE318 pKa = 4.0AEE320 pKa = 3.79FDD322 pKa = 3.69GSGAAADD329 pKa = 4.29TDD331 pKa = 4.35TSCDD335 pKa = 3.31AADD338 pKa = 4.15ASNSGAEE345 pKa = 4.21CHH347 pKa = 5.88GAAPEE352 pKa = 4.14GVEE355 pKa = 4.43GEE357 pKa = 4.62EE358 pKa = 4.44EE359 pKa = 4.29VQQDD363 pKa = 4.03EE364 pKa = 4.58QEE366 pKa = 4.31HH367 pKa = 6.14CAGAGDD373 pKa = 4.28EE374 pKa = 4.2EE375 pKa = 5.01LEE377 pKa = 4.46AVDD380 pKa = 5.35SEE382 pKa = 4.58DD383 pKa = 3.55AAHH386 pKa = 6.17VKK388 pKa = 10.25HH389 pKa = 6.93DD390 pKa = 4.3EE391 pKa = 4.15EE392 pKa = 4.33EE393 pKa = 4.22ALPGGDD399 pKa = 3.92LRR401 pKa = 11.84CTEE404 pKa = 4.21GADD407 pKa = 5.0AAFDD411 pKa = 3.64WAGEE415 pKa = 4.09SDD417 pKa = 3.78VPEE420 pKa = 4.58IFDD423 pKa = 3.39EE424 pKa = 4.83HH425 pKa = 6.48GAAACLGADD434 pKa = 4.82SSWQQQSCGGYY445 pKa = 10.56AVDD448 pKa = 5.15APAGEE453 pKa = 4.87DD454 pKa = 3.73SQDD457 pKa = 3.44SAAEE461 pKa = 3.99EE462 pKa = 4.09QPQQHH467 pKa = 6.29NSTTEE472 pKa = 3.81SDD474 pKa = 3.29GHH476 pKa = 7.47DD477 pKa = 4.18DD478 pKa = 5.15FDD480 pKa = 6.55ALMDD484 pKa = 4.44DD485 pKa = 5.75AIAEE489 pKa = 4.31YY490 pKa = 10.77DD491 pKa = 3.21SDD493 pKa = 5.35AYY495 pKa = 10.72DD496 pKa = 3.32GQYY499 pKa = 11.3DD500 pKa = 3.44VDD502 pKa = 3.5IPYY505 pKa = 10.45DD506 pKa = 3.76AEE508 pKa = 4.53CPAGTAAAVASDD520 pKa = 3.46GGAANRR526 pKa = 11.84ASDD529 pKa = 4.61DD530 pKa = 3.86DD531 pKa = 3.9VAEE534 pKa = 5.26AGLQDD539 pKa = 4.9PLRR542 pKa = 11.84QPPVGMVLLADD553 pKa = 4.04LHH555 pKa = 6.62PCLGSNTSGVSPTALLGSKK574 pKa = 9.96RR575 pKa = 11.84ARR577 pKa = 11.84DD578 pKa = 3.47EE579 pKa = 4.81DD580 pKa = 4.28GEE582 pKa = 4.42EE583 pKa = 4.27ACNAAAPHH591 pKa = 6.12CRR593 pKa = 11.84MRR595 pKa = 11.84PAGFACSALEE605 pKa = 4.04TVQVGSCEE613 pKa = 3.93LL614 pKa = 3.47
Molecular weight: 63.82 kDa
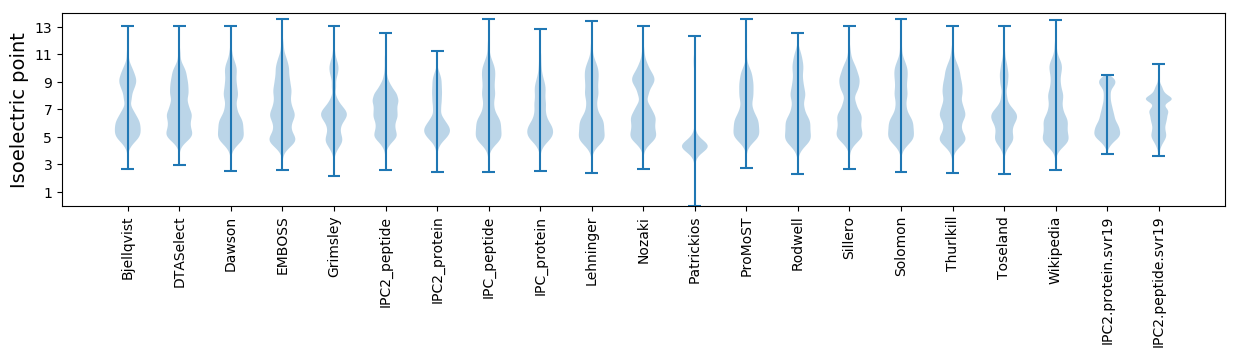
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A150GNU4|A0A150GNU4_GONPE Uncharacterized protein OS=Gonium pectorale OX=33097 GN=GPECTOR_12g418 PE=4 SV=1
MM1 pKa = 7.58AATASMVALVTSMASVAALMAVAALVTAALVTAALVMAALVTAALVTVALVSAALVAAALVTAALMTAALVTAALVTAALMTAALVTAALVTAALVTAALVTAALVTAALVSAALVTAALVTAALGTAALVMAALVMAALVTAALVTAALVTPALVKK158 pKa = 10.49AALVTPAALVTVVVAPVVAAAASVAALVVARR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84WRR193 pKa = 11.84RR194 pKa = 11.84WWRR197 pKa = 11.84RR198 pKa = 11.84PRR200 pKa = 11.84WWPRR204 pKa = 11.84WWPRR208 pKa = 11.84RR209 pKa = 11.84WRR211 pKa = 3.63
MM1 pKa = 7.58AATASMVALVTSMASVAALMAVAALVTAALVTAALVMAALVTAALVTVALVSAALVAAALVTAALMTAALVTAALVTAALMTAALVTAALVTAALVTAALVTAALVTAALVSAALVTAALVTAALGTAALVMAALVMAALVTAALVTAALVTPALVKK158 pKa = 10.49AALVTPAALVTVVVAPVVAAAASVAALVVARR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84WRR193 pKa = 11.84RR194 pKa = 11.84WWRR197 pKa = 11.84RR198 pKa = 11.84PRR200 pKa = 11.84WWPRR204 pKa = 11.84WWPRR208 pKa = 11.84RR209 pKa = 11.84WRR211 pKa = 3.63
Molecular weight: 20.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8125833 |
54 |
8881 |
500.9 |
52.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.459 ± 0.051 | 1.462 ± 0.01 |
4.791 ± 0.013 | 5.552 ± 0.025 |
2.402 ± 0.013 | 10.339 ± 0.029 |
2.039 ± 0.008 | 2.369 ± 0.014 |
2.783 ± 0.019 | 9.655 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.979 ± 0.008 | 2.115 ± 0.014 |
6.855 ± 0.024 | 3.94 ± 0.019 |
6.696 ± 0.019 | 7.064 ± 0.018 |
4.615 ± 0.015 | 6.586 ± 0.018 |
1.302 ± 0.007 | 1.996 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
