
Cryphonectria parasitica mycoreovirus 1 (strain 9B21) (CpMYRV-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Mycoreovirus; Mycoreovirus 1
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
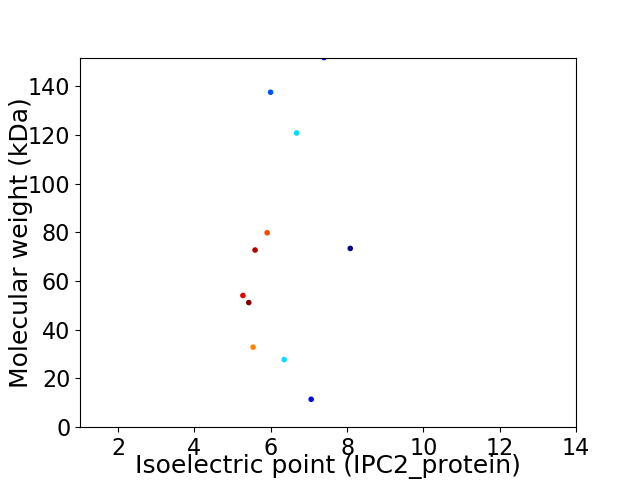
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q65YU9|VP7_MYRV9 Uncharacterized protein VP7 OS=Cryphonectria parasitica mycoreovirus 1 (strain 9B21) OX=230407 GN=S7 PE=4 SV=1
MM1 pKa = 7.92ADD3 pKa = 3.37LTSTSVNQTVLATQPLTTRR22 pKa = 11.84EE23 pKa = 3.97LQQIRR28 pKa = 11.84SMVQTSSNLMQFGNHH43 pKa = 4.96TALPDD48 pKa = 3.6SYY50 pKa = 11.75LEE52 pKa = 3.98TFLFNNGVVYY62 pKa = 10.71EE63 pKa = 4.95DD64 pKa = 4.15GDD66 pKa = 3.79VFYY69 pKa = 10.7AWSEE73 pKa = 4.51TVHH76 pKa = 6.69HH77 pKa = 6.37PTFLDD82 pKa = 3.83AQDD85 pKa = 3.46QRR87 pKa = 11.84YY88 pKa = 9.55SLEE91 pKa = 3.92LHH93 pKa = 6.82PIEE96 pKa = 5.54HH97 pKa = 7.22DD98 pKa = 3.32QTVSPNVSHH107 pKa = 6.49SVWWQCADD115 pKa = 3.39FNKK118 pKa = 9.67FRR120 pKa = 11.84TLSVFGRR127 pKa = 11.84RR128 pKa = 11.84VVPRR132 pKa = 11.84VRR134 pKa = 11.84YY135 pKa = 9.12HH136 pKa = 6.88VDD138 pKa = 2.62EE139 pKa = 4.34FTITFSVSVPLRR151 pKa = 11.84CRR153 pKa = 11.84SFQDD157 pKa = 3.55YY158 pKa = 11.24YY159 pKa = 11.2DD160 pKa = 3.21IHH162 pKa = 7.96VQMKK166 pKa = 8.74MRR168 pKa = 11.84TPSPSVHH175 pKa = 7.02DD176 pKa = 3.8PTHH179 pKa = 6.4ISLASEE185 pKa = 4.16IEE187 pKa = 4.47SISNDD192 pKa = 2.88NDD194 pKa = 3.18VSVPAVVSQPNEE206 pKa = 3.28ITINVGALTPYY217 pKa = 9.91VGHH220 pKa = 6.3SSKK223 pKa = 10.83LAGAALSMIMNIPRR237 pKa = 11.84KK238 pKa = 8.96PVPASPAPRR247 pKa = 11.84VHH249 pKa = 6.43PAVLLTHH256 pKa = 6.51SFFDD260 pKa = 4.04FVRR263 pKa = 11.84EE264 pKa = 4.01QISRR268 pKa = 11.84MDD270 pKa = 3.52ATIMPIHH277 pKa = 6.48LVEE280 pKa = 4.06QTFPEE285 pKa = 4.41AFLAIPVPRR294 pKa = 11.84DD295 pKa = 2.98IEE297 pKa = 4.43TGLVTALRR305 pKa = 11.84SVAMVPTSQYY315 pKa = 9.52NLHH318 pKa = 6.42VMAEE322 pKa = 4.35RR323 pKa = 11.84VQSQSMDD330 pKa = 2.73WTGGNFSRR338 pKa = 11.84IPMHH342 pKa = 7.49DD343 pKa = 4.27DD344 pKa = 4.29DD345 pKa = 7.43IIMTHH350 pKa = 5.96DD351 pKa = 4.53TITALSTGVSTYY363 pKa = 10.01HH364 pKa = 5.76HH365 pKa = 6.94HH366 pKa = 7.23LAITEE371 pKa = 4.15DD372 pKa = 3.64DD373 pKa = 3.57MAVIKK378 pKa = 10.85SRR380 pKa = 11.84VPGVIKK386 pKa = 10.74YY387 pKa = 10.11RR388 pKa = 11.84GSIDD392 pKa = 3.9DD393 pKa = 5.53LKK395 pKa = 11.4ALSNLLEE402 pKa = 4.34SPRR405 pKa = 11.84THH407 pKa = 5.89QVLLHH412 pKa = 6.52SIATIHH418 pKa = 6.84IYY420 pKa = 10.55DD421 pKa = 4.14IGDD424 pKa = 3.31EE425 pKa = 4.35SEE427 pKa = 4.92IDD429 pKa = 3.69DD430 pKa = 4.24QEE432 pKa = 4.36MYY434 pKa = 10.5SKK436 pKa = 10.77KK437 pKa = 10.25ISFLFLLAYY446 pKa = 10.85LMEE449 pKa = 5.03CVTLPTTLSQGFEE462 pKa = 3.99PRR464 pKa = 11.84LPLLPSSRR472 pKa = 11.84VPFYY476 pKa = 11.02LAFGVV481 pKa = 3.62
MM1 pKa = 7.92ADD3 pKa = 3.37LTSTSVNQTVLATQPLTTRR22 pKa = 11.84EE23 pKa = 3.97LQQIRR28 pKa = 11.84SMVQTSSNLMQFGNHH43 pKa = 4.96TALPDD48 pKa = 3.6SYY50 pKa = 11.75LEE52 pKa = 3.98TFLFNNGVVYY62 pKa = 10.71EE63 pKa = 4.95DD64 pKa = 4.15GDD66 pKa = 3.79VFYY69 pKa = 10.7AWSEE73 pKa = 4.51TVHH76 pKa = 6.69HH77 pKa = 6.37PTFLDD82 pKa = 3.83AQDD85 pKa = 3.46QRR87 pKa = 11.84YY88 pKa = 9.55SLEE91 pKa = 3.92LHH93 pKa = 6.82PIEE96 pKa = 5.54HH97 pKa = 7.22DD98 pKa = 3.32QTVSPNVSHH107 pKa = 6.49SVWWQCADD115 pKa = 3.39FNKK118 pKa = 9.67FRR120 pKa = 11.84TLSVFGRR127 pKa = 11.84RR128 pKa = 11.84VVPRR132 pKa = 11.84VRR134 pKa = 11.84YY135 pKa = 9.12HH136 pKa = 6.88VDD138 pKa = 2.62EE139 pKa = 4.34FTITFSVSVPLRR151 pKa = 11.84CRR153 pKa = 11.84SFQDD157 pKa = 3.55YY158 pKa = 11.24YY159 pKa = 11.2DD160 pKa = 3.21IHH162 pKa = 7.96VQMKK166 pKa = 8.74MRR168 pKa = 11.84TPSPSVHH175 pKa = 7.02DD176 pKa = 3.8PTHH179 pKa = 6.4ISLASEE185 pKa = 4.16IEE187 pKa = 4.47SISNDD192 pKa = 2.88NDD194 pKa = 3.18VSVPAVVSQPNEE206 pKa = 3.28ITINVGALTPYY217 pKa = 9.91VGHH220 pKa = 6.3SSKK223 pKa = 10.83LAGAALSMIMNIPRR237 pKa = 11.84KK238 pKa = 8.96PVPASPAPRR247 pKa = 11.84VHH249 pKa = 6.43PAVLLTHH256 pKa = 6.51SFFDD260 pKa = 4.04FVRR263 pKa = 11.84EE264 pKa = 4.01QISRR268 pKa = 11.84MDD270 pKa = 3.52ATIMPIHH277 pKa = 6.48LVEE280 pKa = 4.06QTFPEE285 pKa = 4.41AFLAIPVPRR294 pKa = 11.84DD295 pKa = 2.98IEE297 pKa = 4.43TGLVTALRR305 pKa = 11.84SVAMVPTSQYY315 pKa = 9.52NLHH318 pKa = 6.42VMAEE322 pKa = 4.35RR323 pKa = 11.84VQSQSMDD330 pKa = 2.73WTGGNFSRR338 pKa = 11.84IPMHH342 pKa = 7.49DD343 pKa = 4.27DD344 pKa = 4.29DD345 pKa = 7.43IIMTHH350 pKa = 5.96DD351 pKa = 4.53TITALSTGVSTYY363 pKa = 10.01HH364 pKa = 5.76HH365 pKa = 6.94HH366 pKa = 7.23LAITEE371 pKa = 4.15DD372 pKa = 3.64DD373 pKa = 3.57MAVIKK378 pKa = 10.85SRR380 pKa = 11.84VPGVIKK386 pKa = 10.74YY387 pKa = 10.11RR388 pKa = 11.84GSIDD392 pKa = 3.9DD393 pKa = 5.53LKK395 pKa = 11.4ALSNLLEE402 pKa = 4.34SPRR405 pKa = 11.84THH407 pKa = 5.89QVLLHH412 pKa = 6.52SIATIHH418 pKa = 6.84IYY420 pKa = 10.55DD421 pKa = 4.14IGDD424 pKa = 3.31EE425 pKa = 4.35SEE427 pKa = 4.92IDD429 pKa = 3.69DD430 pKa = 4.24QEE432 pKa = 4.36MYY434 pKa = 10.5SKK436 pKa = 10.77KK437 pKa = 10.25ISFLFLLAYY446 pKa = 10.85LMEE449 pKa = 5.03CVTLPTTLSQGFEE462 pKa = 3.99PRR464 pKa = 11.84LPLLPSSRR472 pKa = 11.84VPFYY476 pKa = 11.02LAFGVV481 pKa = 3.62
Molecular weight: 54.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q65YV1|VP5_MYRV9 Uncharacterized protein VP5 OS=Cryphonectria parasitica mycoreovirus 1 (strain 9B21) OX=230407 GN=S5 PE=4 SV=1
MM1 pKa = 7.61EE2 pKa = 5.75LPHH5 pKa = 6.84DD6 pKa = 4.25KK7 pKa = 10.78AWAYY11 pKa = 11.28LLLTPPPLRR20 pKa = 11.84DD21 pKa = 3.6YY22 pKa = 10.6FIQLPKK28 pKa = 10.27PATLQLALNRR38 pKa = 11.84FAAYY42 pKa = 10.21LEE44 pKa = 4.06KK45 pKa = 10.66QYY47 pKa = 11.29LRR49 pKa = 11.84DD50 pKa = 3.41SLYY53 pKa = 9.81MEE55 pKa = 5.38KK56 pKa = 10.03IVIGTHH62 pKa = 4.35YY63 pKa = 10.36NVRR66 pKa = 11.84KK67 pKa = 10.02NRR69 pKa = 11.84VLQPRR74 pKa = 11.84IFVSPSNYY82 pKa = 8.67TRR84 pKa = 11.84ILSTFYY90 pKa = 11.06DD91 pKa = 4.1HH92 pKa = 7.93PDD94 pKa = 4.06LIAEE98 pKa = 4.21VLPIQHH104 pKa = 7.06LSKK107 pKa = 10.84SQLKK111 pKa = 10.57RR112 pKa = 11.84GGLTKK117 pKa = 10.68NVLTSGLPEE126 pKa = 4.18MNNEE130 pKa = 3.91QNPYY134 pKa = 11.07AVFEE138 pKa = 4.61VNTIDD143 pKa = 5.1DD144 pKa = 4.59LKK146 pKa = 11.33ACFAFPTEE154 pKa = 3.99WLIDD158 pKa = 3.32ILTGRR163 pKa = 11.84NSISLHH169 pKa = 3.96YY170 pKa = 9.61TIDD173 pKa = 3.35RR174 pKa = 11.84TSIPKK179 pKa = 9.48WYY181 pKa = 9.98AHH183 pKa = 6.52LEE185 pKa = 3.85ARR187 pKa = 11.84FQYY190 pKa = 10.91SDD192 pKa = 3.67GVTKK196 pKa = 10.81SLTDD200 pKa = 3.32ADD202 pKa = 4.39LRR204 pKa = 11.84QHH206 pKa = 6.51YY207 pKa = 10.13IGVQPIPVTCEE218 pKa = 3.4EE219 pKa = 5.04LLQKK223 pKa = 10.51MNDD226 pKa = 3.07ASINGNTMYY235 pKa = 10.28RR236 pKa = 11.84VHH238 pKa = 7.84GYY240 pKa = 10.19EE241 pKa = 4.51SIVSSRR247 pKa = 11.84MEE249 pKa = 4.12TVNDD253 pKa = 3.55LVKK256 pKa = 11.01ASGKK260 pKa = 9.83PYY262 pKa = 10.81DD263 pKa = 3.92VVNVQTMSSYY273 pKa = 11.04SRR275 pKa = 11.84SISKK279 pKa = 10.48AINQAVTKK287 pKa = 10.19DD288 pKa = 3.26RR289 pKa = 11.84HH290 pKa = 5.48NKK292 pKa = 9.12HH293 pKa = 6.1VGSTKK298 pKa = 10.48LKK300 pKa = 9.47NSQGPRR306 pKa = 11.84TTIFVMPPYY315 pKa = 9.09ITVDD319 pKa = 3.74VVCSPMTIKK328 pKa = 10.66PGLGPIQTKK337 pKa = 9.16LLKK340 pKa = 10.66SIVDD344 pKa = 3.14QSPRR348 pKa = 11.84RR349 pKa = 11.84FRR351 pKa = 11.84IAYY354 pKa = 7.55AAKK357 pKa = 9.19PQIEE361 pKa = 3.87QDD363 pKa = 3.83VLNKK367 pKa = 10.36RR368 pKa = 11.84IFSTDD373 pKa = 3.44EE374 pKa = 4.08GPEE377 pKa = 4.19SYY379 pKa = 10.48FQGVEE384 pKa = 4.03LLTPEE389 pKa = 4.45AKK391 pKa = 10.25SLLTALPQLVGTFHH405 pKa = 6.57YY406 pKa = 9.77EE407 pKa = 3.83KK408 pKa = 10.45LSYY411 pKa = 11.07AHH413 pKa = 6.42FPSYY417 pKa = 10.97RR418 pKa = 11.84LTAGGYY424 pKa = 8.91NVDD427 pKa = 3.56YY428 pKa = 10.14FKK430 pKa = 11.03RR431 pKa = 11.84GVTVIVARR439 pKa = 11.84KK440 pKa = 9.02HH441 pKa = 5.39GGKK444 pKa = 10.76GMVAKK449 pKa = 9.88LIWKK453 pKa = 9.66LGTPVIDD460 pKa = 3.38SDD462 pKa = 4.32DD463 pKa = 3.48YY464 pKa = 11.73GRR466 pKa = 11.84IIKK469 pKa = 10.13LVEE472 pKa = 4.52AGLTIDD478 pKa = 5.14DD479 pKa = 4.76AVTQLFSMDD488 pKa = 3.18YY489 pKa = 9.86DD490 pKa = 3.67QRR492 pKa = 11.84NSTVSAFDD500 pKa = 3.15EE501 pKa = 4.37HH502 pKa = 7.97MEE504 pKa = 4.28MIVSQSKK511 pKa = 8.4MKK513 pKa = 10.73QEE515 pKa = 3.41ITYY518 pKa = 9.39PRR520 pKa = 11.84ASDD523 pKa = 3.44PRR525 pKa = 11.84ITAFADD531 pKa = 4.47YY532 pKa = 10.13YY533 pKa = 11.07SKK535 pKa = 10.86WFLKK539 pKa = 10.58VPYY542 pKa = 8.89SDD544 pKa = 4.18YY545 pKa = 11.02VASVKK550 pKa = 10.8NFVTKK555 pKa = 10.79NGLSGPNGIQSTNHH569 pKa = 6.2YY570 pKa = 7.41DD571 pKa = 3.41TLVIQVHH578 pKa = 5.63TLPEE582 pKa = 4.03ATQFLGVNNIIEE594 pKa = 4.59VYY596 pKa = 10.12PIIDD600 pKa = 3.85SYY602 pKa = 11.65IGMILRR608 pKa = 11.84QQTSNTCAEE617 pKa = 4.4LLLARR622 pKa = 11.84YY623 pKa = 9.35YY624 pKa = 10.96EE625 pKa = 4.76SIHH628 pKa = 6.39LRR630 pKa = 11.84NFDD633 pKa = 3.93MLPVGVVASTLEE645 pKa = 3.98ALVGG649 pKa = 3.66
MM1 pKa = 7.61EE2 pKa = 5.75LPHH5 pKa = 6.84DD6 pKa = 4.25KK7 pKa = 10.78AWAYY11 pKa = 11.28LLLTPPPLRR20 pKa = 11.84DD21 pKa = 3.6YY22 pKa = 10.6FIQLPKK28 pKa = 10.27PATLQLALNRR38 pKa = 11.84FAAYY42 pKa = 10.21LEE44 pKa = 4.06KK45 pKa = 10.66QYY47 pKa = 11.29LRR49 pKa = 11.84DD50 pKa = 3.41SLYY53 pKa = 9.81MEE55 pKa = 5.38KK56 pKa = 10.03IVIGTHH62 pKa = 4.35YY63 pKa = 10.36NVRR66 pKa = 11.84KK67 pKa = 10.02NRR69 pKa = 11.84VLQPRR74 pKa = 11.84IFVSPSNYY82 pKa = 8.67TRR84 pKa = 11.84ILSTFYY90 pKa = 11.06DD91 pKa = 4.1HH92 pKa = 7.93PDD94 pKa = 4.06LIAEE98 pKa = 4.21VLPIQHH104 pKa = 7.06LSKK107 pKa = 10.84SQLKK111 pKa = 10.57RR112 pKa = 11.84GGLTKK117 pKa = 10.68NVLTSGLPEE126 pKa = 4.18MNNEE130 pKa = 3.91QNPYY134 pKa = 11.07AVFEE138 pKa = 4.61VNTIDD143 pKa = 5.1DD144 pKa = 4.59LKK146 pKa = 11.33ACFAFPTEE154 pKa = 3.99WLIDD158 pKa = 3.32ILTGRR163 pKa = 11.84NSISLHH169 pKa = 3.96YY170 pKa = 9.61TIDD173 pKa = 3.35RR174 pKa = 11.84TSIPKK179 pKa = 9.48WYY181 pKa = 9.98AHH183 pKa = 6.52LEE185 pKa = 3.85ARR187 pKa = 11.84FQYY190 pKa = 10.91SDD192 pKa = 3.67GVTKK196 pKa = 10.81SLTDD200 pKa = 3.32ADD202 pKa = 4.39LRR204 pKa = 11.84QHH206 pKa = 6.51YY207 pKa = 10.13IGVQPIPVTCEE218 pKa = 3.4EE219 pKa = 5.04LLQKK223 pKa = 10.51MNDD226 pKa = 3.07ASINGNTMYY235 pKa = 10.28RR236 pKa = 11.84VHH238 pKa = 7.84GYY240 pKa = 10.19EE241 pKa = 4.51SIVSSRR247 pKa = 11.84MEE249 pKa = 4.12TVNDD253 pKa = 3.55LVKK256 pKa = 11.01ASGKK260 pKa = 9.83PYY262 pKa = 10.81DD263 pKa = 3.92VVNVQTMSSYY273 pKa = 11.04SRR275 pKa = 11.84SISKK279 pKa = 10.48AINQAVTKK287 pKa = 10.19DD288 pKa = 3.26RR289 pKa = 11.84HH290 pKa = 5.48NKK292 pKa = 9.12HH293 pKa = 6.1VGSTKK298 pKa = 10.48LKK300 pKa = 9.47NSQGPRR306 pKa = 11.84TTIFVMPPYY315 pKa = 9.09ITVDD319 pKa = 3.74VVCSPMTIKK328 pKa = 10.66PGLGPIQTKK337 pKa = 9.16LLKK340 pKa = 10.66SIVDD344 pKa = 3.14QSPRR348 pKa = 11.84RR349 pKa = 11.84FRR351 pKa = 11.84IAYY354 pKa = 7.55AAKK357 pKa = 9.19PQIEE361 pKa = 3.87QDD363 pKa = 3.83VLNKK367 pKa = 10.36RR368 pKa = 11.84IFSTDD373 pKa = 3.44EE374 pKa = 4.08GPEE377 pKa = 4.19SYY379 pKa = 10.48FQGVEE384 pKa = 4.03LLTPEE389 pKa = 4.45AKK391 pKa = 10.25SLLTALPQLVGTFHH405 pKa = 6.57YY406 pKa = 9.77EE407 pKa = 3.83KK408 pKa = 10.45LSYY411 pKa = 11.07AHH413 pKa = 6.42FPSYY417 pKa = 10.97RR418 pKa = 11.84LTAGGYY424 pKa = 8.91NVDD427 pKa = 3.56YY428 pKa = 10.14FKK430 pKa = 11.03RR431 pKa = 11.84GVTVIVARR439 pKa = 11.84KK440 pKa = 9.02HH441 pKa = 5.39GGKK444 pKa = 10.76GMVAKK449 pKa = 9.88LIWKK453 pKa = 9.66LGTPVIDD460 pKa = 3.38SDD462 pKa = 4.32DD463 pKa = 3.48YY464 pKa = 11.73GRR466 pKa = 11.84IIKK469 pKa = 10.13LVEE472 pKa = 4.52AGLTIDD478 pKa = 5.14DD479 pKa = 4.76AVTQLFSMDD488 pKa = 3.18YY489 pKa = 9.86DD490 pKa = 3.67QRR492 pKa = 11.84NSTVSAFDD500 pKa = 3.15EE501 pKa = 4.37HH502 pKa = 7.97MEE504 pKa = 4.28MIVSQSKK511 pKa = 8.4MKK513 pKa = 10.73QEE515 pKa = 3.41ITYY518 pKa = 9.39PRR520 pKa = 11.84ASDD523 pKa = 3.44PRR525 pKa = 11.84ITAFADD531 pKa = 4.47YY532 pKa = 10.13YY533 pKa = 11.07SKK535 pKa = 10.86WFLKK539 pKa = 10.58VPYY542 pKa = 8.89SDD544 pKa = 4.18YY545 pKa = 11.02VASVKK550 pKa = 10.8NFVTKK555 pKa = 10.79NGLSGPNGIQSTNHH569 pKa = 6.2YY570 pKa = 7.41DD571 pKa = 3.41TLVIQVHH578 pKa = 5.63TLPEE582 pKa = 4.03ATQFLGVNNIIEE594 pKa = 4.59VYY596 pKa = 10.12PIIDD600 pKa = 3.85SYY602 pKa = 11.65IGMILRR608 pKa = 11.84QQTSNTCAEE617 pKa = 4.4LLLARR622 pKa = 11.84YY623 pKa = 9.35YY624 pKa = 10.96EE625 pKa = 4.76SIHH628 pKa = 6.39LRR630 pKa = 11.84NFDD633 pKa = 3.93MLPVGVVASTLEE645 pKa = 3.98ALVGG649 pKa = 3.66
Molecular weight: 73.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7268 |
101 |
1354 |
660.7 |
73.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.302 ± 0.154 | 1.238 ± 0.22 |
5.407 ± 0.198 | 3.949 ± 0.161 |
4.513 ± 0.274 | 4.898 ± 0.384 |
3.261 ± 0.197 | 6.976 ± 0.284 |
3.082 ± 0.431 | 9.439 ± 0.309 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.655 ± 0.27 | 4.444 ± 0.205 |
6.288 ± 0.399 | 3.605 ± 0.343 |
5.476 ± 0.381 | 8.847 ± 0.336 |
7.843 ± 0.293 | 6.962 ± 0.413 |
0.757 ± 0.075 | 4.059 ± 0.296 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
