
Naegleria fowleri (Brain eating amoeba)
Taxonomy: cellular organisms; Eukaryota; Discoba; Heterolobosea; Tetramitia; Eutetramitia; Vahlkampfiidae; Naegleria
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
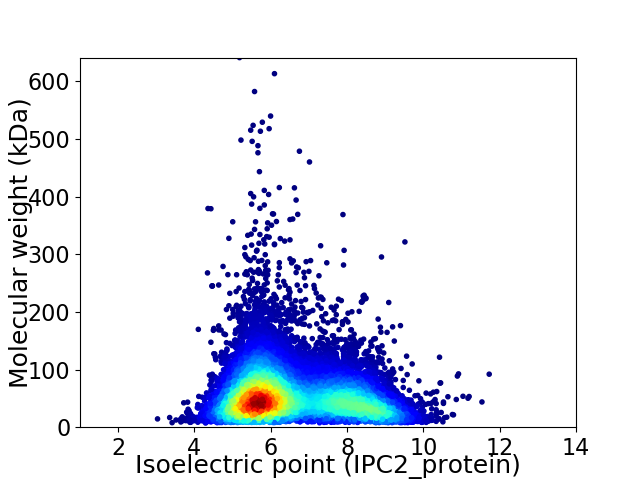
Virtual 2D-PAGE plot for 13596 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A5AU86|A0A6A5AU86_NAEFO RING-type domain-containing protein OS=Naegleria fowleri OX=5763 GN=FDP41_010076 PE=4 SV=1
MM1 pKa = 7.75DD2 pKa = 4.38LWYY5 pKa = 10.38CEE7 pKa = 4.33KK8 pKa = 10.45EE9 pKa = 3.96IEE11 pKa = 4.53SADD14 pKa = 4.31RR15 pKa = 11.84MIEE18 pKa = 3.73QWKK21 pKa = 8.21TEE23 pKa = 3.76IGEE26 pKa = 4.31GFDD29 pKa = 3.53VNFMRR34 pKa = 11.84FEE36 pKa = 4.34LLSEE40 pKa = 4.21SLKK43 pKa = 10.7HH44 pKa = 5.61IQRR47 pKa = 11.84EE48 pKa = 4.3DD49 pKa = 3.23EE50 pKa = 4.84FLNLSNNGYY59 pKa = 10.3CDD61 pKa = 4.95DD62 pKa = 6.25DD63 pKa = 4.4EE64 pKa = 5.04EE65 pKa = 6.52CDD67 pKa = 5.05DD68 pKa = 5.44EE69 pKa = 4.91EE70 pKa = 4.52EE71 pKa = 4.41CDD73 pKa = 3.84YY74 pKa = 11.74EE75 pKa = 4.28EE76 pKa = 4.61YY77 pKa = 10.47EE78 pKa = 4.2YY79 pKa = 11.42SRR81 pKa = 11.84DD82 pKa = 3.55CATPISPAACEE93 pKa = 3.89YY94 pKa = 10.52FSFVSEE100 pKa = 4.47CSDD103 pKa = 5.12DD104 pKa = 3.87EE105 pKa = 6.56DD106 pKa = 4.05EE107 pKa = 5.5DD108 pKa = 4.42EE109 pKa = 4.9YY110 pKa = 11.98YY111 pKa = 10.94SDD113 pKa = 6.08DD114 pKa = 4.12EE115 pKa = 5.85DD116 pKa = 5.66DD117 pKa = 4.78EE118 pKa = 5.15EE119 pKa = 6.01DD120 pKa = 5.33FEE122 pKa = 5.87EE123 pKa = 4.9EE124 pKa = 4.49DD125 pKa = 3.36NCVYY129 pKa = 11.46DD130 pKa = 4.68NMDD133 pKa = 3.94TVSVICDD140 pKa = 3.36QNHH143 pKa = 6.57PNGNSIEE150 pKa = 4.29DD151 pKa = 3.69TTADD155 pKa = 3.52NSEE158 pKa = 4.58KK159 pKa = 10.66GHH161 pKa = 5.72PHH163 pKa = 5.49QDD165 pKa = 2.8ASNKK169 pKa = 9.28HH170 pKa = 5.44MKK172 pKa = 9.17TFSRR176 pKa = 11.84DD177 pKa = 3.58LIHH180 pKa = 7.67DD181 pKa = 4.31EE182 pKa = 4.41YY183 pKa = 10.94IGGLLHH189 pKa = 7.15EE190 pKa = 5.71FGPCSTHH197 pKa = 7.53SNTASNKK204 pKa = 9.98LQMDD208 pKa = 3.67DD209 pKa = 4.05HH210 pKa = 7.18EE211 pKa = 4.99EE212 pKa = 4.27GQEE215 pKa = 4.02EE216 pKa = 4.92EE217 pKa = 4.55PSSPQPIMISNCDD230 pKa = 3.48TLSEE234 pKa = 4.27QEE236 pKa = 3.95MSSINSSDD244 pKa = 3.44NEE246 pKa = 4.33TSHH249 pKa = 7.41SCIVNNKK256 pKa = 5.24QTVDD260 pKa = 3.98FSKK263 pKa = 9.61HH264 pKa = 3.57QQVIFEE270 pKa = 4.25
MM1 pKa = 7.75DD2 pKa = 4.38LWYY5 pKa = 10.38CEE7 pKa = 4.33KK8 pKa = 10.45EE9 pKa = 3.96IEE11 pKa = 4.53SADD14 pKa = 4.31RR15 pKa = 11.84MIEE18 pKa = 3.73QWKK21 pKa = 8.21TEE23 pKa = 3.76IGEE26 pKa = 4.31GFDD29 pKa = 3.53VNFMRR34 pKa = 11.84FEE36 pKa = 4.34LLSEE40 pKa = 4.21SLKK43 pKa = 10.7HH44 pKa = 5.61IQRR47 pKa = 11.84EE48 pKa = 4.3DD49 pKa = 3.23EE50 pKa = 4.84FLNLSNNGYY59 pKa = 10.3CDD61 pKa = 4.95DD62 pKa = 6.25DD63 pKa = 4.4EE64 pKa = 5.04EE65 pKa = 6.52CDD67 pKa = 5.05DD68 pKa = 5.44EE69 pKa = 4.91EE70 pKa = 4.52EE71 pKa = 4.41CDD73 pKa = 3.84YY74 pKa = 11.74EE75 pKa = 4.28EE76 pKa = 4.61YY77 pKa = 10.47EE78 pKa = 4.2YY79 pKa = 11.42SRR81 pKa = 11.84DD82 pKa = 3.55CATPISPAACEE93 pKa = 3.89YY94 pKa = 10.52FSFVSEE100 pKa = 4.47CSDD103 pKa = 5.12DD104 pKa = 3.87EE105 pKa = 6.56DD106 pKa = 4.05EE107 pKa = 5.5DD108 pKa = 4.42EE109 pKa = 4.9YY110 pKa = 11.98YY111 pKa = 10.94SDD113 pKa = 6.08DD114 pKa = 4.12EE115 pKa = 5.85DD116 pKa = 5.66DD117 pKa = 4.78EE118 pKa = 5.15EE119 pKa = 6.01DD120 pKa = 5.33FEE122 pKa = 5.87EE123 pKa = 4.9EE124 pKa = 4.49DD125 pKa = 3.36NCVYY129 pKa = 11.46DD130 pKa = 4.68NMDD133 pKa = 3.94TVSVICDD140 pKa = 3.36QNHH143 pKa = 6.57PNGNSIEE150 pKa = 4.29DD151 pKa = 3.69TTADD155 pKa = 3.52NSEE158 pKa = 4.58KK159 pKa = 10.66GHH161 pKa = 5.72PHH163 pKa = 5.49QDD165 pKa = 2.8ASNKK169 pKa = 9.28HH170 pKa = 5.44MKK172 pKa = 9.17TFSRR176 pKa = 11.84DD177 pKa = 3.58LIHH180 pKa = 7.67DD181 pKa = 4.31EE182 pKa = 4.41YY183 pKa = 10.94IGGLLHH189 pKa = 7.15EE190 pKa = 5.71FGPCSTHH197 pKa = 7.53SNTASNKK204 pKa = 9.98LQMDD208 pKa = 3.67DD209 pKa = 4.05HH210 pKa = 7.18EE211 pKa = 4.99EE212 pKa = 4.27GQEE215 pKa = 4.02EE216 pKa = 4.92EE217 pKa = 4.55PSSPQPIMISNCDD230 pKa = 3.48TLSEE234 pKa = 4.27QEE236 pKa = 3.95MSSINSSDD244 pKa = 3.44NEE246 pKa = 4.33TSHH249 pKa = 7.41SCIVNNKK256 pKa = 5.24QTVDD260 pKa = 3.98FSKK263 pKa = 9.61HH264 pKa = 3.57QQVIFEE270 pKa = 4.25
Molecular weight: 31.22 kDa
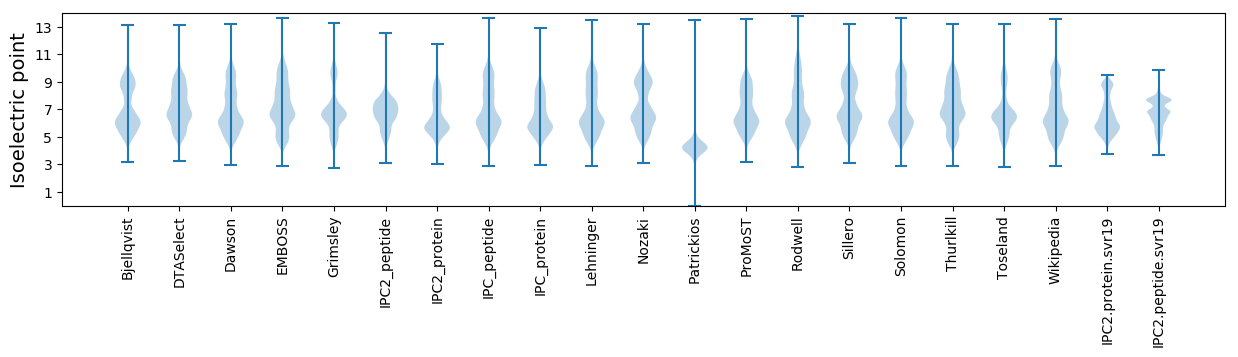
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A5C3D5|A0A6A5C3D5_NAEFO Guanylate cyclase domain-containing protein OS=Naegleria fowleri OX=5763 GN=FDP41_000812 PE=4 SV=1
MM1 pKa = 7.49NKK3 pKa = 10.1FILLALAMLFCMVMFVATTTTQAASADD30 pKa = 3.74TLAASTLCFGLKK42 pKa = 9.42VKK44 pKa = 10.57RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84ATCIRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.7RR56 pKa = 11.84KK57 pKa = 8.64LLRR60 pKa = 11.84RR61 pKa = 11.84VPSKK65 pKa = 9.96RR66 pKa = 11.84RR67 pKa = 11.84AVIRR71 pKa = 11.84RR72 pKa = 11.84LLKK75 pKa = 8.81RR76 pKa = 11.84CRR78 pKa = 11.84RR79 pKa = 11.84NRR81 pKa = 11.84RR82 pKa = 11.84AVKK85 pKa = 10.14KK86 pKa = 10.06ALKK89 pKa = 9.97RR90 pKa = 11.84SRR92 pKa = 11.84KK93 pKa = 7.13LARR96 pKa = 11.84RR97 pKa = 11.84IRR99 pKa = 11.84RR100 pKa = 11.84VKK102 pKa = 10.12SKK104 pKa = 10.0RR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84AIRR111 pKa = 11.84RR112 pKa = 11.84ALKK115 pKa = 9.89RR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84QAKK122 pKa = 9.54KK123 pKa = 9.46IRR125 pKa = 11.84RR126 pKa = 11.84AARR129 pKa = 11.84RR130 pKa = 11.84CVKK133 pKa = 10.22RR134 pKa = 11.84ASKK137 pKa = 10.31KK138 pKa = 9.26ARR140 pKa = 11.84KK141 pKa = 8.91ARR143 pKa = 11.84KK144 pKa = 8.0LARR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84ARR151 pKa = 11.84RR152 pKa = 11.84AARR155 pKa = 11.84KK156 pKa = 8.44ALKK159 pKa = 9.77RR160 pKa = 11.84ARR162 pKa = 11.84RR163 pKa = 11.84LAKK166 pKa = 9.61RR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84ALRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AALKK178 pKa = 10.17RR179 pKa = 11.84RR180 pKa = 11.84AALRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84ARR188 pKa = 11.84RR189 pKa = 11.84LSKK192 pKa = 10.41RR193 pKa = 11.84SSLAKK198 pKa = 8.56RR199 pKa = 11.84CRR201 pKa = 11.84RR202 pKa = 11.84FVRR205 pKa = 11.84RR206 pKa = 11.84ATKK209 pKa = 10.35KK210 pKa = 10.08AVKK213 pKa = 10.04KK214 pKa = 9.4ATKK217 pKa = 10.2KK218 pKa = 9.96LQKK221 pKa = 10.08KK222 pKa = 8.87LKK224 pKa = 9.36KK225 pKa = 9.78VKK227 pKa = 10.05KK228 pKa = 8.95AAKK231 pKa = 9.77KK232 pKa = 9.88AVKK235 pKa = 9.62KK236 pKa = 10.58AKK238 pKa = 10.15KK239 pKa = 8.87QVKK242 pKa = 8.93KK243 pKa = 10.6AKK245 pKa = 9.8KK246 pKa = 8.92AARR249 pKa = 11.84KK250 pKa = 9.17AKK252 pKa = 9.57RR253 pKa = 11.84AAKK256 pKa = 9.96RR257 pKa = 11.84RR258 pKa = 11.84VALRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84APRR268 pKa = 11.84KK269 pKa = 8.52CGRR272 pKa = 11.84ARR274 pKa = 11.84RR275 pKa = 11.84STRR278 pKa = 11.84KK279 pKa = 8.3ATKK282 pKa = 9.91KK283 pKa = 9.7AAKK286 pKa = 9.82KK287 pKa = 8.79ATKK290 pKa = 9.91KK291 pKa = 9.66QLKK294 pKa = 9.4KK295 pKa = 10.3QLKK298 pKa = 9.58KK299 pKa = 10.55LQKK302 pKa = 9.94KK303 pKa = 9.27LKK305 pKa = 8.88KK306 pKa = 9.98AKK308 pKa = 10.14KK309 pKa = 9.4AVKK312 pKa = 9.59KK313 pKa = 10.54AKK315 pKa = 10.03KK316 pKa = 8.44QAKK319 pKa = 8.52KK320 pKa = 10.46AKK322 pKa = 9.77KK323 pKa = 9.24AAKK326 pKa = 9.22KK327 pKa = 10.02AKK329 pKa = 9.8KK330 pKa = 9.35AAKK333 pKa = 9.55KK334 pKa = 9.15RR335 pKa = 11.84AARR338 pKa = 11.84RR339 pKa = 11.84KK340 pKa = 9.49VVLKK344 pKa = 10.71KK345 pKa = 10.4KK346 pKa = 9.87AAKK349 pKa = 9.73KK350 pKa = 9.9LNKK353 pKa = 9.31IVKK356 pKa = 9.88KK357 pKa = 10.41LLRR360 pKa = 11.84LKK362 pKa = 10.58KK363 pKa = 10.48KK364 pKa = 9.95LGKK367 pKa = 10.08RR368 pKa = 11.84FNPKK372 pKa = 9.14KK373 pKa = 9.92HH374 pKa = 5.47VPKK377 pKa = 10.47KK378 pKa = 9.66VRR380 pKa = 11.84KK381 pKa = 9.13LLRR384 pKa = 11.84KK385 pKa = 9.97LKK387 pKa = 9.25IQKK390 pKa = 8.9RR391 pKa = 11.84CRR393 pKa = 11.84RR394 pKa = 11.84SRR396 pKa = 11.84RR397 pKa = 11.84ARR399 pKa = 11.84RR400 pKa = 11.84TRR402 pKa = 11.84RR403 pKa = 11.84CRR405 pKa = 11.84KK406 pKa = 9.63VAVKK410 pKa = 10.42LPKK413 pKa = 10.35NLLKK417 pKa = 10.8KK418 pKa = 10.27LNKK421 pKa = 8.73VQKK424 pKa = 10.72SNN426 pKa = 3.27
MM1 pKa = 7.49NKK3 pKa = 10.1FILLALAMLFCMVMFVATTTTQAASADD30 pKa = 3.74TLAASTLCFGLKK42 pKa = 9.42VKK44 pKa = 10.57RR45 pKa = 11.84NRR47 pKa = 11.84RR48 pKa = 11.84ATCIRR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 5.7RR56 pKa = 11.84KK57 pKa = 8.64LLRR60 pKa = 11.84RR61 pKa = 11.84VPSKK65 pKa = 9.96RR66 pKa = 11.84RR67 pKa = 11.84AVIRR71 pKa = 11.84RR72 pKa = 11.84LLKK75 pKa = 8.81RR76 pKa = 11.84CRR78 pKa = 11.84RR79 pKa = 11.84NRR81 pKa = 11.84RR82 pKa = 11.84AVKK85 pKa = 10.14KK86 pKa = 10.06ALKK89 pKa = 9.97RR90 pKa = 11.84SRR92 pKa = 11.84KK93 pKa = 7.13LARR96 pKa = 11.84RR97 pKa = 11.84IRR99 pKa = 11.84RR100 pKa = 11.84VKK102 pKa = 10.12SKK104 pKa = 10.0RR105 pKa = 11.84ARR107 pKa = 11.84RR108 pKa = 11.84AIRR111 pKa = 11.84RR112 pKa = 11.84ALKK115 pKa = 9.89RR116 pKa = 11.84ARR118 pKa = 11.84RR119 pKa = 11.84QAKK122 pKa = 9.54KK123 pKa = 9.46IRR125 pKa = 11.84RR126 pKa = 11.84AARR129 pKa = 11.84RR130 pKa = 11.84CVKK133 pKa = 10.22RR134 pKa = 11.84ASKK137 pKa = 10.31KK138 pKa = 9.26ARR140 pKa = 11.84KK141 pKa = 8.91ARR143 pKa = 11.84KK144 pKa = 8.0LARR147 pKa = 11.84RR148 pKa = 11.84RR149 pKa = 11.84ARR151 pKa = 11.84RR152 pKa = 11.84AARR155 pKa = 11.84KK156 pKa = 8.44ALKK159 pKa = 9.77RR160 pKa = 11.84ARR162 pKa = 11.84RR163 pKa = 11.84LAKK166 pKa = 9.61RR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84ALRR172 pKa = 11.84RR173 pKa = 11.84RR174 pKa = 11.84AALKK178 pKa = 10.17RR179 pKa = 11.84RR180 pKa = 11.84AALRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84ARR188 pKa = 11.84RR189 pKa = 11.84LSKK192 pKa = 10.41RR193 pKa = 11.84SSLAKK198 pKa = 8.56RR199 pKa = 11.84CRR201 pKa = 11.84RR202 pKa = 11.84FVRR205 pKa = 11.84RR206 pKa = 11.84ATKK209 pKa = 10.35KK210 pKa = 10.08AVKK213 pKa = 10.04KK214 pKa = 9.4ATKK217 pKa = 10.2KK218 pKa = 9.96LQKK221 pKa = 10.08KK222 pKa = 8.87LKK224 pKa = 9.36KK225 pKa = 9.78VKK227 pKa = 10.05KK228 pKa = 8.95AAKK231 pKa = 9.77KK232 pKa = 9.88AVKK235 pKa = 9.62KK236 pKa = 10.58AKK238 pKa = 10.15KK239 pKa = 8.87QVKK242 pKa = 8.93KK243 pKa = 10.6AKK245 pKa = 9.8KK246 pKa = 8.92AARR249 pKa = 11.84KK250 pKa = 9.17AKK252 pKa = 9.57RR253 pKa = 11.84AAKK256 pKa = 9.96RR257 pKa = 11.84RR258 pKa = 11.84VALRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84APRR268 pKa = 11.84KK269 pKa = 8.52CGRR272 pKa = 11.84ARR274 pKa = 11.84RR275 pKa = 11.84STRR278 pKa = 11.84KK279 pKa = 8.3ATKK282 pKa = 9.91KK283 pKa = 9.7AAKK286 pKa = 9.82KK287 pKa = 8.79ATKK290 pKa = 9.91KK291 pKa = 9.66QLKK294 pKa = 9.4KK295 pKa = 10.3QLKK298 pKa = 9.58KK299 pKa = 10.55LQKK302 pKa = 9.94KK303 pKa = 9.27LKK305 pKa = 8.88KK306 pKa = 9.98AKK308 pKa = 10.14KK309 pKa = 9.4AVKK312 pKa = 9.59KK313 pKa = 10.54AKK315 pKa = 10.03KK316 pKa = 8.44QAKK319 pKa = 8.52KK320 pKa = 10.46AKK322 pKa = 9.77KK323 pKa = 9.24AAKK326 pKa = 9.22KK327 pKa = 10.02AKK329 pKa = 9.8KK330 pKa = 9.35AAKK333 pKa = 9.55KK334 pKa = 9.15RR335 pKa = 11.84AARR338 pKa = 11.84RR339 pKa = 11.84KK340 pKa = 9.49VVLKK344 pKa = 10.71KK345 pKa = 10.4KK346 pKa = 9.87AAKK349 pKa = 9.73KK350 pKa = 9.9LNKK353 pKa = 9.31IVKK356 pKa = 9.88KK357 pKa = 10.41LLRR360 pKa = 11.84LKK362 pKa = 10.58KK363 pKa = 10.48KK364 pKa = 9.95LGKK367 pKa = 10.08RR368 pKa = 11.84FNPKK372 pKa = 9.14KK373 pKa = 9.92HH374 pKa = 5.47VPKK377 pKa = 10.47KK378 pKa = 9.66VRR380 pKa = 11.84KK381 pKa = 9.13LLRR384 pKa = 11.84KK385 pKa = 9.97LKK387 pKa = 9.25IQKK390 pKa = 8.9RR391 pKa = 11.84CRR393 pKa = 11.84RR394 pKa = 11.84SRR396 pKa = 11.84RR397 pKa = 11.84ARR399 pKa = 11.84RR400 pKa = 11.84TRR402 pKa = 11.84RR403 pKa = 11.84CRR405 pKa = 11.84KK406 pKa = 9.63VAVKK410 pKa = 10.42LPKK413 pKa = 10.35NLLKK417 pKa = 10.8KK418 pKa = 10.27LNKK421 pKa = 8.73VQKK424 pKa = 10.72SNN426 pKa = 3.27
Molecular weight: 50.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7262538 |
66 |
5561 |
534.2 |
60.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.529 ± 0.016 | 1.592 ± 0.012 |
5.106 ± 0.012 | 6.775 ± 0.023 |
4.61 ± 0.015 | 4.414 ± 0.026 |
2.79 ± 0.014 | 6.363 ± 0.017 |
7.246 ± 0.024 | 9.264 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.008 | 6.217 ± 0.016 |
4.082 ± 0.017 | 4.709 ± 0.016 |
4.001 ± 0.012 | 9.947 ± 0.029 |
6.347 ± 0.018 | 5.423 ± 0.015 |
0.813 ± 0.005 | 3.318 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
