
Daedalea quercina L-15889
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Fomitopsidaceae; Daedalea; Daedalea quercina
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
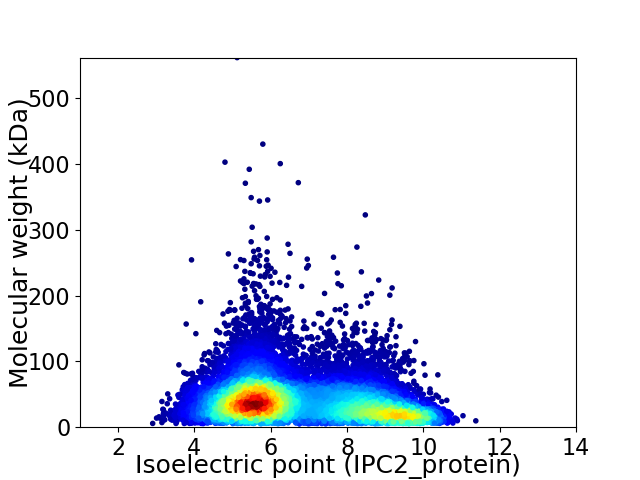
Virtual 2D-PAGE plot for 12170 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165LHK6|A0A165LHK6_9APHY Glutathione transferase OS=Daedalea quercina L-15889 OX=1314783 GN=DAEQUDRAFT_732662 PE=3 SV=1
MM1 pKa = 6.94NTFFGLLIASLCGLTNALYY20 pKa = 9.97IPVRR24 pKa = 11.84VNHH27 pKa = 6.94DD28 pKa = 3.44AQASSSGTGVHH39 pKa = 6.02NPAIRR44 pKa = 11.84LVGSFRR50 pKa = 11.84QQSDD54 pKa = 3.3DD55 pKa = 3.76ANNGSVVNFQDD66 pKa = 3.15TVYY69 pKa = 9.36ATNMTIGGQDD79 pKa = 3.47VLIQLDD85 pKa = 3.98TGSSDD90 pKa = 2.79LWVNLDD96 pKa = 3.25EE97 pKa = 5.71GEE99 pKa = 4.42IEE101 pKa = 4.62FSNTTDD107 pKa = 3.79LPAQEE112 pKa = 5.2AYY114 pKa = 10.52GIGQIAGTIQFANVEE129 pKa = 4.4LGDD132 pKa = 4.04NMVPNQAFMNVSNATNFGTIFNDD155 pKa = 4.02GVRR158 pKa = 11.84GILGLAFDD166 pKa = 4.9AGSTIEE172 pKa = 4.09STITEE177 pKa = 4.46AYY179 pKa = 10.5GEE181 pKa = 4.12DD182 pKa = 3.95SEE184 pKa = 5.71LGEE187 pKa = 4.37SFLTNVFQQDD197 pKa = 3.05TSKK200 pKa = 10.98PLFTVQLGRR209 pKa = 11.84ADD211 pKa = 3.86DD212 pKa = 4.05PQYY215 pKa = 8.32TTEE218 pKa = 4.01GAFTIGEE225 pKa = 4.27YY226 pKa = 10.85LPDD229 pKa = 3.72YY230 pKa = 8.57EE231 pKa = 5.17TIEE234 pKa = 4.78DD235 pKa = 3.83MPKK238 pKa = 10.57LEE240 pKa = 4.4RR241 pKa = 11.84FPNNGTGAAPRR252 pKa = 11.84WSTVMSGMTVNGEE265 pKa = 3.68PFQFNTSGVPGTPEE279 pKa = 3.89GSVVAVLDD287 pKa = 3.59TGFTFPPIPKK297 pKa = 8.86PAVDD301 pKa = 5.13AIYY304 pKa = 10.64GGIDD308 pKa = 3.11GAYY311 pKa = 9.63YY312 pKa = 8.59DD313 pKa = 4.71TNSSLWIVPCNKK325 pKa = 9.11ATTLEE330 pKa = 4.24FQFGEE335 pKa = 4.3LSVPIHH341 pKa = 6.45PLDD344 pKa = 3.49LTTIVQINNTQVCVNTFRR362 pKa = 11.84PSTFPVNTQFDD373 pKa = 4.78LVLGDD378 pKa = 4.55AFLKK382 pKa = 10.56NVYY385 pKa = 10.7ASFNYY390 pKa = 10.44GNMSEE395 pKa = 4.77DD396 pKa = 3.33AVPYY400 pKa = 8.99MQLLPTTDD408 pKa = 2.65MHH410 pKa = 6.64AASKK414 pKa = 10.65EE415 pKa = 3.87FDD417 pKa = 3.65SVRR420 pKa = 11.84SAMYY424 pKa = 10.06AQAAASAAASSASASAASAYY444 pKa = 9.9AALVSGSASTACSTAMPTATPLFASAWSSSAPSAVMTDD482 pKa = 3.27STPAIIASPDD492 pKa = 3.31LSGLGNGNSSNPTAASSANGSSATDD517 pKa = 3.4ASSTDD522 pKa = 3.56PAATDD527 pKa = 3.45ASSTDD532 pKa = 3.4GSSATATSDD541 pKa = 2.79TSTPTSTDD549 pKa = 3.18DD550 pKa = 3.59SSASTTSDD558 pKa = 2.79TSASSPTDD566 pKa = 3.12SATSSTATSTDD577 pKa = 2.84SSTDD581 pKa = 3.32PSATSTDD588 pKa = 3.24SSTDD592 pKa = 3.32PSATSTDD599 pKa = 3.24SSTDD603 pKa = 3.27PSATSTASAADD614 pKa = 3.86PNATSTDD621 pKa = 3.5SSSDD625 pKa = 3.41TAGTTSTASAADD637 pKa = 3.77PSATSSGDD645 pKa = 3.25SSSGGNSSSTGSLPIDD661 pKa = 4.08GSSSSDD667 pKa = 3.41DD668 pKa = 3.58GSGSLQEE675 pKa = 4.22NTSLLDD681 pKa = 3.34NSGGSKK687 pKa = 10.3QSTFGNSLYY696 pKa = 11.08GDD698 pKa = 4.19GGNGYY703 pKa = 10.32ARR705 pKa = 11.84AFRR708 pKa = 11.84RR709 pKa = 11.84SYY711 pKa = 10.93RR712 pKa = 11.84EE713 pKa = 3.6PTKK716 pKa = 10.24TVHH719 pKa = 7.44DD720 pKa = 4.78CLTPMIEE727 pKa = 4.37EE728 pKa = 4.09YY729 pKa = 10.93GALLFALIAGTFIISFALCLVTVVLAIRR757 pKa = 11.84NHH759 pKa = 5.61SRR761 pKa = 11.84SQARR765 pKa = 11.84KK766 pKa = 7.39RR767 pKa = 11.84AEE769 pKa = 4.05YY770 pKa = 10.05FVLEE774 pKa = 4.31ANAEE778 pKa = 4.07AEE780 pKa = 4.38ADD782 pKa = 3.76PEE784 pKa = 4.29AACTATYY791 pKa = 9.88HH792 pKa = 6.56YY793 pKa = 10.92SDD795 pKa = 5.47DD796 pKa = 3.97YY797 pKa = 12.02
MM1 pKa = 6.94NTFFGLLIASLCGLTNALYY20 pKa = 9.97IPVRR24 pKa = 11.84VNHH27 pKa = 6.94DD28 pKa = 3.44AQASSSGTGVHH39 pKa = 6.02NPAIRR44 pKa = 11.84LVGSFRR50 pKa = 11.84QQSDD54 pKa = 3.3DD55 pKa = 3.76ANNGSVVNFQDD66 pKa = 3.15TVYY69 pKa = 9.36ATNMTIGGQDD79 pKa = 3.47VLIQLDD85 pKa = 3.98TGSSDD90 pKa = 2.79LWVNLDD96 pKa = 3.25EE97 pKa = 5.71GEE99 pKa = 4.42IEE101 pKa = 4.62FSNTTDD107 pKa = 3.79LPAQEE112 pKa = 5.2AYY114 pKa = 10.52GIGQIAGTIQFANVEE129 pKa = 4.4LGDD132 pKa = 4.04NMVPNQAFMNVSNATNFGTIFNDD155 pKa = 4.02GVRR158 pKa = 11.84GILGLAFDD166 pKa = 4.9AGSTIEE172 pKa = 4.09STITEE177 pKa = 4.46AYY179 pKa = 10.5GEE181 pKa = 4.12DD182 pKa = 3.95SEE184 pKa = 5.71LGEE187 pKa = 4.37SFLTNVFQQDD197 pKa = 3.05TSKK200 pKa = 10.98PLFTVQLGRR209 pKa = 11.84ADD211 pKa = 3.86DD212 pKa = 4.05PQYY215 pKa = 8.32TTEE218 pKa = 4.01GAFTIGEE225 pKa = 4.27YY226 pKa = 10.85LPDD229 pKa = 3.72YY230 pKa = 8.57EE231 pKa = 5.17TIEE234 pKa = 4.78DD235 pKa = 3.83MPKK238 pKa = 10.57LEE240 pKa = 4.4RR241 pKa = 11.84FPNNGTGAAPRR252 pKa = 11.84WSTVMSGMTVNGEE265 pKa = 3.68PFQFNTSGVPGTPEE279 pKa = 3.89GSVVAVLDD287 pKa = 3.59TGFTFPPIPKK297 pKa = 8.86PAVDD301 pKa = 5.13AIYY304 pKa = 10.64GGIDD308 pKa = 3.11GAYY311 pKa = 9.63YY312 pKa = 8.59DD313 pKa = 4.71TNSSLWIVPCNKK325 pKa = 9.11ATTLEE330 pKa = 4.24FQFGEE335 pKa = 4.3LSVPIHH341 pKa = 6.45PLDD344 pKa = 3.49LTTIVQINNTQVCVNTFRR362 pKa = 11.84PSTFPVNTQFDD373 pKa = 4.78LVLGDD378 pKa = 4.55AFLKK382 pKa = 10.56NVYY385 pKa = 10.7ASFNYY390 pKa = 10.44GNMSEE395 pKa = 4.77DD396 pKa = 3.33AVPYY400 pKa = 8.99MQLLPTTDD408 pKa = 2.65MHH410 pKa = 6.64AASKK414 pKa = 10.65EE415 pKa = 3.87FDD417 pKa = 3.65SVRR420 pKa = 11.84SAMYY424 pKa = 10.06AQAAASAAASSASASAASAYY444 pKa = 9.9AALVSGSASTACSTAMPTATPLFASAWSSSAPSAVMTDD482 pKa = 3.27STPAIIASPDD492 pKa = 3.31LSGLGNGNSSNPTAASSANGSSATDD517 pKa = 3.4ASSTDD522 pKa = 3.56PAATDD527 pKa = 3.45ASSTDD532 pKa = 3.4GSSATATSDD541 pKa = 2.79TSTPTSTDD549 pKa = 3.18DD550 pKa = 3.59SSASTTSDD558 pKa = 2.79TSASSPTDD566 pKa = 3.12SATSSTATSTDD577 pKa = 2.84SSTDD581 pKa = 3.32PSATSTDD588 pKa = 3.24SSTDD592 pKa = 3.32PSATSTDD599 pKa = 3.24SSTDD603 pKa = 3.27PSATSTASAADD614 pKa = 3.86PNATSTDD621 pKa = 3.5SSSDD625 pKa = 3.41TAGTTSTASAADD637 pKa = 3.77PSATSSGDD645 pKa = 3.25SSSGGNSSSTGSLPIDD661 pKa = 4.08GSSSSDD667 pKa = 3.41DD668 pKa = 3.58GSGSLQEE675 pKa = 4.22NTSLLDD681 pKa = 3.34NSGGSKK687 pKa = 10.3QSTFGNSLYY696 pKa = 11.08GDD698 pKa = 4.19GGNGYY703 pKa = 10.32ARR705 pKa = 11.84AFRR708 pKa = 11.84RR709 pKa = 11.84SYY711 pKa = 10.93RR712 pKa = 11.84EE713 pKa = 3.6PTKK716 pKa = 10.24TVHH719 pKa = 7.44DD720 pKa = 4.78CLTPMIEE727 pKa = 4.37EE728 pKa = 4.09YY729 pKa = 10.93GALLFALIAGTFIISFALCLVTVVLAIRR757 pKa = 11.84NHH759 pKa = 5.61SRR761 pKa = 11.84SQARR765 pKa = 11.84KK766 pKa = 7.39RR767 pKa = 11.84AEE769 pKa = 4.05YY770 pKa = 10.05FVLEE774 pKa = 4.31ANAEE778 pKa = 4.07AEE780 pKa = 4.38ADD782 pKa = 3.76PEE784 pKa = 4.29AACTATYY791 pKa = 9.88HH792 pKa = 6.56YY793 pKa = 10.92SDD795 pKa = 5.47DD796 pKa = 3.97YY797 pKa = 12.02
Molecular weight: 82.11 kDa
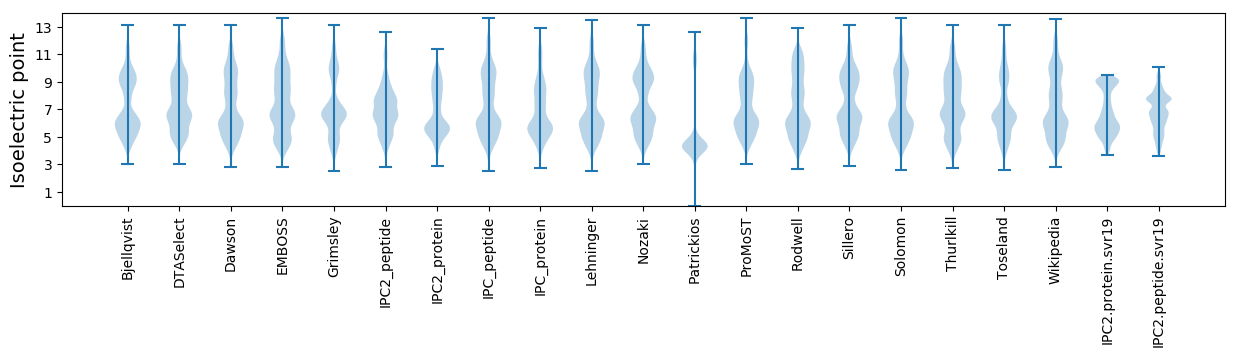
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165THV8|A0A165THV8_9APHY Uncharacterized protein OS=Daedalea quercina L-15889 OX=1314783 GN=DAEQUDRAFT_721516 PE=4 SV=1
PP1 pKa = 6.32SARR4 pKa = 11.84RR5 pKa = 11.84GRR7 pKa = 11.84AASPTKK13 pKa = 8.96PTSTIAPTKK22 pKa = 9.38RR23 pKa = 11.84TAPPVKK29 pKa = 9.93RR30 pKa = 11.84ATSPTKK36 pKa = 10.42RR37 pKa = 11.84ATSPAKK43 pKa = 10.18RR44 pKa = 11.84AASPIKK50 pKa = 10.28RR51 pKa = 11.84AAPPVKK57 pKa = 10.03RR58 pKa = 11.84AASPVKK64 pKa = 10.21RR65 pKa = 11.84AASPVKK71 pKa = 10.35RR72 pKa = 11.84AGSPVKK78 pKa = 10.32RR79 pKa = 11.84AASPVKK85 pKa = 10.21RR86 pKa = 11.84AASPVKK92 pKa = 10.27RR93 pKa = 11.84AA94 pKa = 3.43
PP1 pKa = 6.32SARR4 pKa = 11.84RR5 pKa = 11.84GRR7 pKa = 11.84AASPTKK13 pKa = 8.96PTSTIAPTKK22 pKa = 9.38RR23 pKa = 11.84TAPPVKK29 pKa = 9.93RR30 pKa = 11.84ATSPTKK36 pKa = 10.42RR37 pKa = 11.84ATSPAKK43 pKa = 10.18RR44 pKa = 11.84AASPIKK50 pKa = 10.28RR51 pKa = 11.84AAPPVKK57 pKa = 10.03RR58 pKa = 11.84AASPVKK64 pKa = 10.21RR65 pKa = 11.84AASPVKK71 pKa = 10.35RR72 pKa = 11.84AGSPVKK78 pKa = 10.32RR79 pKa = 11.84AASPVKK85 pKa = 10.21RR86 pKa = 11.84AASPVKK92 pKa = 10.27RR93 pKa = 11.84AA94 pKa = 3.43
Molecular weight: 9.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4971741 |
49 |
5065 |
408.5 |
45.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.359 ± 0.02 | 1.314 ± 0.009 |
5.558 ± 0.016 | 5.908 ± 0.021 |
3.459 ± 0.014 | 6.629 ± 0.02 |
2.565 ± 0.013 | 4.441 ± 0.014 |
4.081 ± 0.02 | 9.172 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.156 ± 0.009 | 3.012 ± 0.01 |
6.68 ± 0.029 | 3.688 ± 0.014 |
6.769 ± 0.024 | 8.453 ± 0.035 |
5.969 ± 0.017 | 6.621 ± 0.016 |
1.456 ± 0.009 | 2.708 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
