
Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; Clostridium cellulovorans
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
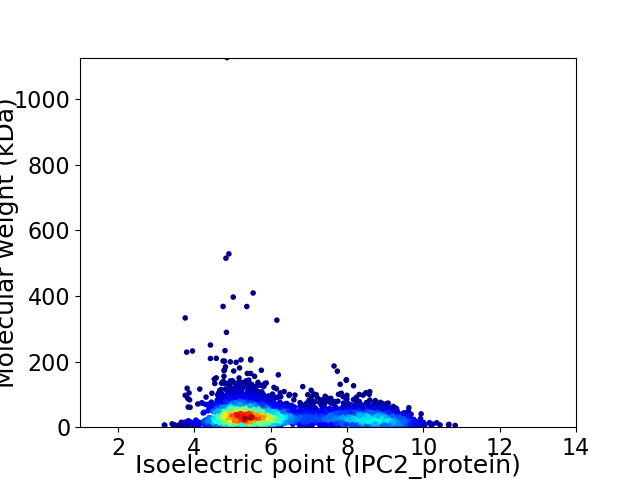
Virtual 2D-PAGE plot for 4211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9SPW3|D9SPW3_CLOC7 Uncharacterized protein OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=Clocel_2383 PE=4 SV=1
MM1 pKa = 7.56ILTYY5 pKa = 9.22SEE7 pKa = 4.95KK8 pKa = 10.96GDD10 pKa = 3.67GQDD13 pKa = 4.48IIVDD17 pKa = 3.54YY18 pKa = 11.08SGEE21 pKa = 4.19ANTIIDD27 pKa = 3.55TYY29 pKa = 10.64IFRR32 pKa = 11.84KK33 pKa = 10.4GYY35 pKa = 8.9GQVNIYY41 pKa = 9.91EE42 pKa = 4.27YY43 pKa = 10.87SGEE46 pKa = 4.24ANTIIDD52 pKa = 3.98TLKK55 pKa = 11.24LEE57 pKa = 5.3DD58 pKa = 4.12INPNEE63 pKa = 4.44ISLNCNRR70 pKa = 11.84YY71 pKa = 9.81DD72 pKa = 5.82LIMKK76 pKa = 10.19LNEE79 pKa = 4.33SKK81 pKa = 11.06DD82 pKa = 3.71SITIQGYY89 pKa = 9.42FSSDD93 pKa = 2.57LHH95 pKa = 8.25KK96 pKa = 10.45IEE98 pKa = 5.57KK99 pKa = 9.92IQFADD104 pKa = 3.76GTVWDD109 pKa = 4.14INTIKK114 pKa = 10.8SSLVYY119 pKa = 10.98SNGTVGIDD127 pKa = 3.27SLNAEE132 pKa = 4.94GEE134 pKa = 4.51VIMNGYY140 pKa = 9.89EE141 pKa = 4.05GDD143 pKa = 3.55DD144 pKa = 3.78TLRR147 pKa = 11.84SGAGNDD153 pKa = 3.68KK154 pKa = 10.69IYY156 pKa = 11.07GGTGNDD162 pKa = 2.88QVYY165 pKa = 10.95GNGGNDD171 pKa = 3.63LLYY174 pKa = 11.3GEE176 pKa = 5.81DD177 pKa = 5.05GDD179 pKa = 4.75DD180 pKa = 4.39KK181 pKa = 11.54LYY183 pKa = 11.24GGNGDD188 pKa = 5.14DD189 pKa = 4.78ILDD192 pKa = 3.97GGEE195 pKa = 4.22GKK197 pKa = 10.29DD198 pKa = 3.54YY199 pKa = 11.16LYY201 pKa = 11.39GEE203 pKa = 4.71NGNDD207 pKa = 4.56LLDD210 pKa = 4.12GKK212 pKa = 10.6NGGGYY217 pKa = 10.26LEE219 pKa = 5.26GGAGVDD225 pKa = 3.21TYY227 pKa = 11.08IFRR230 pKa = 11.84KK231 pKa = 10.39GYY233 pKa = 8.9GQVNIYY239 pKa = 9.91EE240 pKa = 4.27YY241 pKa = 10.87SGEE244 pKa = 4.24ANTIIDD250 pKa = 3.98TLKK253 pKa = 11.24LEE255 pKa = 5.3DD256 pKa = 4.12INPNEE261 pKa = 4.44ISLNCNRR268 pKa = 11.84YY269 pKa = 9.81DD270 pKa = 5.82LIMKK274 pKa = 10.19LNEE277 pKa = 4.33SKK279 pKa = 11.06DD280 pKa = 3.71SITIQGYY287 pKa = 9.42FSSDD291 pKa = 2.57LHH293 pKa = 8.25KK294 pKa = 10.45IEE296 pKa = 5.57KK297 pKa = 9.92IQFADD302 pKa = 3.76GTVWDD307 pKa = 4.14INTIKK312 pKa = 10.8SSLVYY317 pKa = 10.98SNGTVGIDD325 pKa = 3.27SLNAEE330 pKa = 4.94GEE332 pKa = 4.51VIMNGYY338 pKa = 9.89EE339 pKa = 4.05GDD341 pKa = 3.55DD342 pKa = 3.78TLRR345 pKa = 11.84SGAGNDD351 pKa = 3.68KK352 pKa = 10.69IYY354 pKa = 11.07GGTGNDD360 pKa = 2.88QVYY363 pKa = 10.95GNGGNDD369 pKa = 3.63LLYY372 pKa = 11.3GEE374 pKa = 5.81DD375 pKa = 5.05GDD377 pKa = 4.75DD378 pKa = 4.39KK379 pKa = 11.54LYY381 pKa = 11.24GGNGDD386 pKa = 5.14DD387 pKa = 4.78ILDD390 pKa = 3.97GGEE393 pKa = 4.22GKK395 pKa = 10.29DD396 pKa = 3.54YY397 pKa = 11.16LYY399 pKa = 11.39GEE401 pKa = 4.71NGNDD405 pKa = 4.56LLDD408 pKa = 4.12GKK410 pKa = 10.6NGGGYY415 pKa = 10.45LEE417 pKa = 5.18GGAGNDD423 pKa = 3.28TYY425 pKa = 11.04IFSKK429 pKa = 11.08GDD431 pKa = 3.49GQDD434 pKa = 2.95IIYY437 pKa = 10.53NSSSLDD443 pKa = 3.29TDD445 pKa = 3.39NDD447 pKa = 3.87KK448 pKa = 11.58LSLNDD453 pKa = 3.35VNLSEE458 pKa = 4.21TAFIKK463 pKa = 11.02SNTDD467 pKa = 4.01LIIKK471 pKa = 10.2LNEE474 pKa = 4.0TTDD477 pKa = 3.26SVTILGYY484 pKa = 10.45FNYY487 pKa = 10.25GKK489 pKa = 10.8YY490 pKa = 10.19SLEE493 pKa = 4.2EE494 pKa = 3.74IEE496 pKa = 4.76FMNGEE501 pKa = 4.36TLTPEE506 pKa = 4.2DD507 pKa = 4.04VNNVILGKK515 pKa = 10.13YY516 pKa = 8.84VSHH519 pKa = 7.65TIPQYY524 pKa = 11.02NISLDD529 pKa = 3.46KK530 pKa = 10.98AIEE533 pKa = 4.08ILSEE537 pKa = 3.98ASNPSIFNITGTSTSSDD554 pKa = 2.6IMTDD558 pKa = 2.4ILLNMNSS565 pKa = 3.3
MM1 pKa = 7.56ILTYY5 pKa = 9.22SEE7 pKa = 4.95KK8 pKa = 10.96GDD10 pKa = 3.67GQDD13 pKa = 4.48IIVDD17 pKa = 3.54YY18 pKa = 11.08SGEE21 pKa = 4.19ANTIIDD27 pKa = 3.55TYY29 pKa = 10.64IFRR32 pKa = 11.84KK33 pKa = 10.4GYY35 pKa = 8.9GQVNIYY41 pKa = 9.91EE42 pKa = 4.27YY43 pKa = 10.87SGEE46 pKa = 4.24ANTIIDD52 pKa = 3.98TLKK55 pKa = 11.24LEE57 pKa = 5.3DD58 pKa = 4.12INPNEE63 pKa = 4.44ISLNCNRR70 pKa = 11.84YY71 pKa = 9.81DD72 pKa = 5.82LIMKK76 pKa = 10.19LNEE79 pKa = 4.33SKK81 pKa = 11.06DD82 pKa = 3.71SITIQGYY89 pKa = 9.42FSSDD93 pKa = 2.57LHH95 pKa = 8.25KK96 pKa = 10.45IEE98 pKa = 5.57KK99 pKa = 9.92IQFADD104 pKa = 3.76GTVWDD109 pKa = 4.14INTIKK114 pKa = 10.8SSLVYY119 pKa = 10.98SNGTVGIDD127 pKa = 3.27SLNAEE132 pKa = 4.94GEE134 pKa = 4.51VIMNGYY140 pKa = 9.89EE141 pKa = 4.05GDD143 pKa = 3.55DD144 pKa = 3.78TLRR147 pKa = 11.84SGAGNDD153 pKa = 3.68KK154 pKa = 10.69IYY156 pKa = 11.07GGTGNDD162 pKa = 2.88QVYY165 pKa = 10.95GNGGNDD171 pKa = 3.63LLYY174 pKa = 11.3GEE176 pKa = 5.81DD177 pKa = 5.05GDD179 pKa = 4.75DD180 pKa = 4.39KK181 pKa = 11.54LYY183 pKa = 11.24GGNGDD188 pKa = 5.14DD189 pKa = 4.78ILDD192 pKa = 3.97GGEE195 pKa = 4.22GKK197 pKa = 10.29DD198 pKa = 3.54YY199 pKa = 11.16LYY201 pKa = 11.39GEE203 pKa = 4.71NGNDD207 pKa = 4.56LLDD210 pKa = 4.12GKK212 pKa = 10.6NGGGYY217 pKa = 10.26LEE219 pKa = 5.26GGAGVDD225 pKa = 3.21TYY227 pKa = 11.08IFRR230 pKa = 11.84KK231 pKa = 10.39GYY233 pKa = 8.9GQVNIYY239 pKa = 9.91EE240 pKa = 4.27YY241 pKa = 10.87SGEE244 pKa = 4.24ANTIIDD250 pKa = 3.98TLKK253 pKa = 11.24LEE255 pKa = 5.3DD256 pKa = 4.12INPNEE261 pKa = 4.44ISLNCNRR268 pKa = 11.84YY269 pKa = 9.81DD270 pKa = 5.82LIMKK274 pKa = 10.19LNEE277 pKa = 4.33SKK279 pKa = 11.06DD280 pKa = 3.71SITIQGYY287 pKa = 9.42FSSDD291 pKa = 2.57LHH293 pKa = 8.25KK294 pKa = 10.45IEE296 pKa = 5.57KK297 pKa = 9.92IQFADD302 pKa = 3.76GTVWDD307 pKa = 4.14INTIKK312 pKa = 10.8SSLVYY317 pKa = 10.98SNGTVGIDD325 pKa = 3.27SLNAEE330 pKa = 4.94GEE332 pKa = 4.51VIMNGYY338 pKa = 9.89EE339 pKa = 4.05GDD341 pKa = 3.55DD342 pKa = 3.78TLRR345 pKa = 11.84SGAGNDD351 pKa = 3.68KK352 pKa = 10.69IYY354 pKa = 11.07GGTGNDD360 pKa = 2.88QVYY363 pKa = 10.95GNGGNDD369 pKa = 3.63LLYY372 pKa = 11.3GEE374 pKa = 5.81DD375 pKa = 5.05GDD377 pKa = 4.75DD378 pKa = 4.39KK379 pKa = 11.54LYY381 pKa = 11.24GGNGDD386 pKa = 5.14DD387 pKa = 4.78ILDD390 pKa = 3.97GGEE393 pKa = 4.22GKK395 pKa = 10.29DD396 pKa = 3.54YY397 pKa = 11.16LYY399 pKa = 11.39GEE401 pKa = 4.71NGNDD405 pKa = 4.56LLDD408 pKa = 4.12GKK410 pKa = 10.6NGGGYY415 pKa = 10.45LEE417 pKa = 5.18GGAGNDD423 pKa = 3.28TYY425 pKa = 11.04IFSKK429 pKa = 11.08GDD431 pKa = 3.49GQDD434 pKa = 2.95IIYY437 pKa = 10.53NSSSLDD443 pKa = 3.29TDD445 pKa = 3.39NDD447 pKa = 3.87KK448 pKa = 11.58LSLNDD453 pKa = 3.35VNLSEE458 pKa = 4.21TAFIKK463 pKa = 11.02SNTDD467 pKa = 4.01LIIKK471 pKa = 10.2LNEE474 pKa = 4.0TTDD477 pKa = 3.26SVTILGYY484 pKa = 10.45FNYY487 pKa = 10.25GKK489 pKa = 10.8YY490 pKa = 10.19SLEE493 pKa = 4.2EE494 pKa = 3.74IEE496 pKa = 4.76FMNGEE501 pKa = 4.36TLTPEE506 pKa = 4.2DD507 pKa = 4.04VNNVILGKK515 pKa = 10.13YY516 pKa = 8.84VSHH519 pKa = 7.65TIPQYY524 pKa = 11.02NISLDD529 pKa = 3.46KK530 pKa = 10.98AIEE533 pKa = 4.08ILSEE537 pKa = 3.98ASNPSIFNITGTSTSSDD554 pKa = 2.6IMTDD558 pKa = 2.4ILLNMNSS565 pKa = 3.3
Molecular weight: 61.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9SRQ7|D9SRQ7_CLOC7 Uncharacterized protein OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=Clocel_0653 PE=4 SV=1
MM1 pKa = 7.5LKK3 pKa = 10.2SKK5 pKa = 10.87KK6 pKa = 10.42NIIARR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.15IVNLPISKK23 pKa = 9.71FPYY26 pKa = 8.08YY27 pKa = 10.21RR28 pKa = 11.84LYY30 pKa = 10.75RR31 pKa = 11.84PTYY34 pKa = 10.09ALTLPEE40 pKa = 4.0VNRR43 pKa = 11.84LKK45 pKa = 10.72GRR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84TTVPNIVGTITATVGEE66 pKa = 4.71YY67 pKa = 10.66NPNTNRR73 pKa = 11.84VEE75 pKa = 4.19LKK77 pKa = 10.8NIISDD82 pKa = 3.74RR83 pKa = 11.84TGVSYY88 pKa = 11.66GNLSYY93 pKa = 11.26LLDD96 pKa = 5.04EE97 pKa = 4.57IAGLQVLPKK106 pKa = 10.97DD107 pKa = 3.34NGTPGGSKK115 pKa = 10.2DD116 pKa = 3.68CQITGGKK123 pKa = 9.06GKK125 pKa = 10.59YY126 pKa = 7.59ITSGEE131 pKa = 4.12TSIGNVSVKK140 pKa = 8.62YY141 pKa = 8.95TIHH144 pKa = 6.25EE145 pKa = 4.2CMIEE149 pKa = 4.05IIPRR153 pKa = 11.84VDD155 pKa = 3.17GLPPRR160 pKa = 11.84RR161 pKa = 11.84LIINRR166 pKa = 11.84NNTRR170 pKa = 11.84VQTMFRR176 pKa = 11.84TSPRR180 pKa = 11.84NRR182 pKa = 11.84IKK184 pKa = 9.85FTAWIQNNTVWVEE197 pKa = 3.25IEE199 pKa = 3.93YY200 pKa = 10.31QRR202 pKa = 11.84RR203 pKa = 11.84TRR205 pKa = 11.84FGRR208 pKa = 11.84WQRR211 pKa = 11.84INEE214 pKa = 3.86VRR216 pKa = 11.84TKK218 pKa = 10.89LGTWQDD224 pKa = 3.49LKK226 pKa = 11.09II227 pKa = 4.32
MM1 pKa = 7.5LKK3 pKa = 10.2SKK5 pKa = 10.87KK6 pKa = 10.42NIIARR11 pKa = 11.84LRR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 9.15IVNLPISKK23 pKa = 9.71FPYY26 pKa = 8.08YY27 pKa = 10.21RR28 pKa = 11.84LYY30 pKa = 10.75RR31 pKa = 11.84PTYY34 pKa = 10.09ALTLPEE40 pKa = 4.0VNRR43 pKa = 11.84LKK45 pKa = 10.72GRR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84TTVPNIVGTITATVGEE66 pKa = 4.71YY67 pKa = 10.66NPNTNRR73 pKa = 11.84VEE75 pKa = 4.19LKK77 pKa = 10.8NIISDD82 pKa = 3.74RR83 pKa = 11.84TGVSYY88 pKa = 11.66GNLSYY93 pKa = 11.26LLDD96 pKa = 5.04EE97 pKa = 4.57IAGLQVLPKK106 pKa = 10.97DD107 pKa = 3.34NGTPGGSKK115 pKa = 10.2DD116 pKa = 3.68CQITGGKK123 pKa = 9.06GKK125 pKa = 10.59YY126 pKa = 7.59ITSGEE131 pKa = 4.12TSIGNVSVKK140 pKa = 8.62YY141 pKa = 8.95TIHH144 pKa = 6.25EE145 pKa = 4.2CMIEE149 pKa = 4.05IIPRR153 pKa = 11.84VDD155 pKa = 3.17GLPPRR160 pKa = 11.84RR161 pKa = 11.84LIINRR166 pKa = 11.84NNTRR170 pKa = 11.84VQTMFRR176 pKa = 11.84TSPRR180 pKa = 11.84NRR182 pKa = 11.84IKK184 pKa = 9.85FTAWIQNNTVWVEE197 pKa = 3.25IEE199 pKa = 3.93YY200 pKa = 10.31QRR202 pKa = 11.84RR203 pKa = 11.84TRR205 pKa = 11.84FGRR208 pKa = 11.84WQRR211 pKa = 11.84INEE214 pKa = 3.86VRR216 pKa = 11.84TKK218 pKa = 10.89LGTWQDD224 pKa = 3.49LKK226 pKa = 11.09II227 pKa = 4.32
Molecular weight: 26.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1429936 |
30 |
9858 |
339.6 |
38.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.863 ± 0.041 | 1.107 ± 0.014 |
5.72 ± 0.034 | 7.362 ± 0.06 |
4.348 ± 0.031 | 6.356 ± 0.049 |
1.374 ± 0.016 | 9.486 ± 0.048 |
8.465 ± 0.041 | 8.789 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.424 ± 0.018 | 6.273 ± 0.036 |
2.782 ± 0.024 | 2.585 ± 0.019 |
3.304 ± 0.025 | 6.571 ± 0.037 |
5.536 ± 0.046 | 6.531 ± 0.033 |
0.817 ± 0.014 | 4.305 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
