
Symphysodon discus adomavirus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA viruses; Adomaviridae
Average proteome isoelectric point is 7.23
Get precalculated fractions of proteins
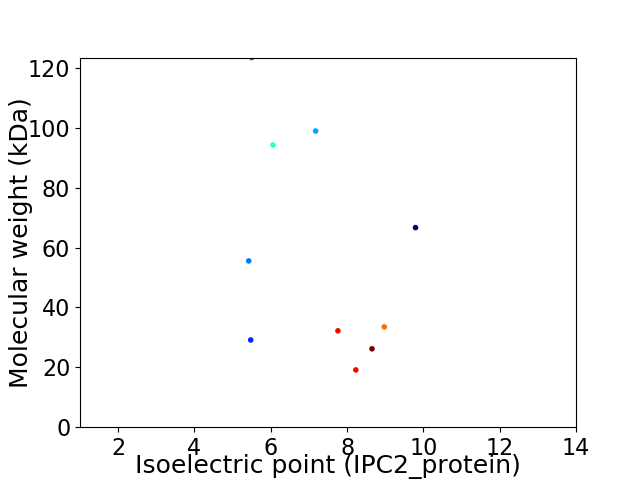
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S1MK48|A0A2S1MK48_9VIRU LO8 OS=Symphysodon discus adomavirus 1 OX=2175118 PE=4 SV=1
MM1 pKa = 6.94ATSRR5 pKa = 11.84QVQSLGRR12 pKa = 11.84TPGGMGYY19 pKa = 10.26SFLGTFDD26 pKa = 6.3DD27 pKa = 4.23NDD29 pKa = 3.43QNNYY33 pKa = 9.74CVLDD37 pKa = 4.38LSAHH41 pKa = 5.84PVACEE46 pKa = 3.41RR47 pKa = 11.84DD48 pKa = 3.61PYY50 pKa = 11.5GNGLAAISVTQITVSNCALHH70 pKa = 6.77IGNHH74 pKa = 6.1LGNHH78 pKa = 6.25YY79 pKa = 10.06IYY81 pKa = 11.03VGGARR86 pKa = 11.84VQLPDD91 pKa = 3.32VSNTSVTQFLDD102 pKa = 3.35YY103 pKa = 11.05LQACNPSLSFEE114 pKa = 4.32FKK116 pKa = 10.54LGRR119 pKa = 11.84LTYY122 pKa = 8.8TGNSAVIIPVRR133 pKa = 11.84TGIGQHH139 pKa = 6.26AKK141 pKa = 10.17PIPHH145 pKa = 7.09NLTHH149 pKa = 6.3MLGFTGLGEE158 pKa = 4.18VEE160 pKa = 4.86YY161 pKa = 10.26EE162 pKa = 3.64NDD164 pKa = 3.32MCVVITPDD172 pKa = 3.22VLGNSAPDD180 pKa = 3.24LFGPVRR186 pKa = 11.84DD187 pKa = 4.26LVVSCAEE194 pKa = 3.75VTYY197 pKa = 10.63NRR199 pKa = 11.84HH200 pKa = 4.94SRR202 pKa = 11.84QSVAVCSPCGTPGQAYY218 pKa = 8.22TRR220 pKa = 11.84DD221 pKa = 3.6CSDD224 pKa = 4.04LVRR227 pKa = 11.84EE228 pKa = 4.12LNIPGGYY235 pKa = 9.71LCSLTFTLTDD245 pKa = 3.38AADD248 pKa = 3.7RR249 pKa = 11.84PVRR252 pKa = 11.84FVHH255 pKa = 6.55GVPLICVRR263 pKa = 11.84ILPITLQQ270 pKa = 3.33
MM1 pKa = 6.94ATSRR5 pKa = 11.84QVQSLGRR12 pKa = 11.84TPGGMGYY19 pKa = 10.26SFLGTFDD26 pKa = 6.3DD27 pKa = 4.23NDD29 pKa = 3.43QNNYY33 pKa = 9.74CVLDD37 pKa = 4.38LSAHH41 pKa = 5.84PVACEE46 pKa = 3.41RR47 pKa = 11.84DD48 pKa = 3.61PYY50 pKa = 11.5GNGLAAISVTQITVSNCALHH70 pKa = 6.77IGNHH74 pKa = 6.1LGNHH78 pKa = 6.25YY79 pKa = 10.06IYY81 pKa = 11.03VGGARR86 pKa = 11.84VQLPDD91 pKa = 3.32VSNTSVTQFLDD102 pKa = 3.35YY103 pKa = 11.05LQACNPSLSFEE114 pKa = 4.32FKK116 pKa = 10.54LGRR119 pKa = 11.84LTYY122 pKa = 8.8TGNSAVIIPVRR133 pKa = 11.84TGIGQHH139 pKa = 6.26AKK141 pKa = 10.17PIPHH145 pKa = 7.09NLTHH149 pKa = 6.3MLGFTGLGEE158 pKa = 4.18VEE160 pKa = 4.86YY161 pKa = 10.26EE162 pKa = 3.64NDD164 pKa = 3.32MCVVITPDD172 pKa = 3.22VLGNSAPDD180 pKa = 3.24LFGPVRR186 pKa = 11.84DD187 pKa = 4.26LVVSCAEE194 pKa = 3.75VTYY197 pKa = 10.63NRR199 pKa = 11.84HH200 pKa = 4.94SRR202 pKa = 11.84QSVAVCSPCGTPGQAYY218 pKa = 8.22TRR220 pKa = 11.84DD221 pKa = 3.6CSDD224 pKa = 4.04LVRR227 pKa = 11.84EE228 pKa = 4.12LNIPGGYY235 pKa = 9.71LCSLTFTLTDD245 pKa = 3.38AADD248 pKa = 3.7RR249 pKa = 11.84PVRR252 pKa = 11.84FVHH255 pKa = 6.55GVPLICVRR263 pKa = 11.84ILPITLQQ270 pKa = 3.33
Molecular weight: 29.13 kDa
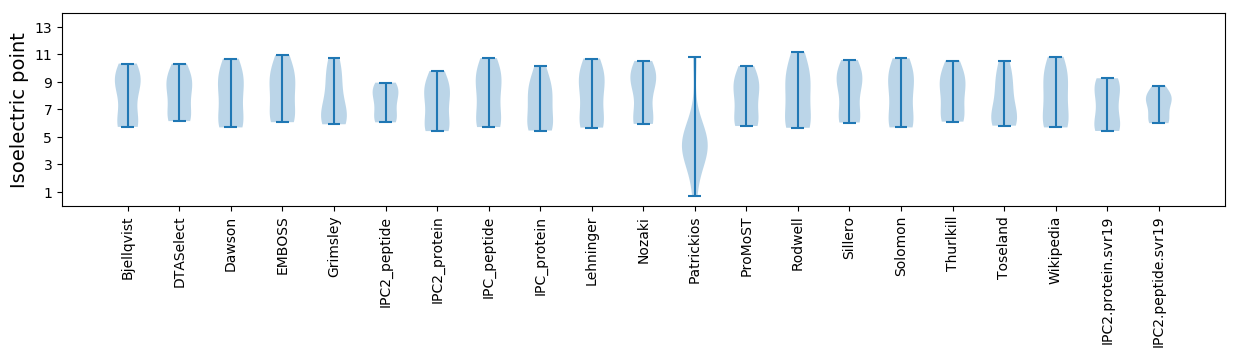
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S1MK68|A0A2S1MK68_9VIRU EO4 OS=Symphysodon discus adomavirus 1 OX=2175118 PE=4 SV=1
MM1 pKa = 7.77AKK3 pKa = 9.01TRR5 pKa = 11.84LLPSKK10 pKa = 10.64KK11 pKa = 9.44KK12 pKa = 10.52GKK14 pKa = 9.69KK15 pKa = 7.4HH16 pKa = 5.39TPNKK20 pKa = 9.94VKK22 pKa = 10.25IKK24 pKa = 10.38KK25 pKa = 8.79EE26 pKa = 3.98PKK28 pKa = 9.99SGITALRR35 pKa = 11.84SYY37 pKa = 11.16KK38 pKa = 10.52HH39 pKa = 5.53FTRR42 pKa = 11.84RR43 pKa = 11.84WNKK46 pKa = 10.25KK47 pKa = 5.85NTQEE51 pKa = 4.29KK52 pKa = 9.9SSPVQTLVKK61 pKa = 10.46VEE63 pKa = 4.34NNIQSSHH70 pKa = 5.73TGGKK74 pKa = 8.78QRR76 pKa = 11.84STMPKK81 pKa = 9.49LRR83 pKa = 11.84AKK85 pKa = 10.21PVVKK89 pKa = 10.26KK90 pKa = 8.89RR91 pKa = 11.84TRR93 pKa = 11.84GSVRR97 pKa = 11.84RR98 pKa = 11.84TPYY101 pKa = 9.43TKK103 pKa = 10.02PNHH106 pKa = 5.82SQASKK111 pKa = 10.5YY112 pKa = 10.58NSTRR116 pKa = 11.84ITKK119 pKa = 7.54STRR122 pKa = 11.84KK123 pKa = 8.3NTTTAPIITVSPNMTFNPQNSNVSSPRR150 pKa = 11.84VSNALVPYY158 pKa = 9.73NHH160 pKa = 6.32NQAQQGLRR168 pKa = 11.84RR169 pKa = 11.84MVEE172 pKa = 4.2KK173 pKa = 10.56EE174 pKa = 3.77SSAPGLASRR183 pKa = 11.84VLRR186 pKa = 11.84RR187 pKa = 11.84MINVALPTLGSAALASMTGIPSFLTNAAVNAVSSAITSDD226 pKa = 3.39EE227 pKa = 4.09DD228 pKa = 3.6VGPVTPTIPIYY239 pKa = 10.76HH240 pKa = 7.49EE241 pKa = 4.56STDD244 pKa = 3.59SAQPALRR251 pKa = 11.84YY252 pKa = 9.72QPIPQYY258 pKa = 11.11SGGPKK263 pKa = 9.72PDD265 pKa = 4.57DD266 pKa = 4.86DD267 pKa = 5.04SPGLLSTAQNLFEE280 pKa = 5.21NMLAYY285 pKa = 10.59VPTLTTLAGAGHH297 pKa = 6.93KK298 pKa = 10.04VYY300 pKa = 10.62QAMRR304 pKa = 11.84GGVAPYY310 pKa = 10.62VSGTHH315 pKa = 6.6TYY317 pKa = 9.2TPQASYY323 pKa = 10.42IQDD326 pKa = 3.33RR327 pKa = 11.84YY328 pKa = 10.3YY329 pKa = 10.92RR330 pKa = 11.84PAPYY334 pKa = 9.62ISSQQYY340 pKa = 9.78RR341 pKa = 11.84GGSGYY346 pKa = 9.96IPSAACRR353 pKa = 11.84VSVHH357 pKa = 5.99NPRR360 pKa = 11.84QYY362 pKa = 11.48ALRR365 pKa = 11.84GTKK368 pKa = 8.13HH369 pKa = 5.6TYY371 pKa = 10.04RR372 pKa = 11.84NRR374 pKa = 11.84AVHH377 pKa = 6.39NSGKK381 pKa = 10.46YY382 pKa = 7.77YY383 pKa = 10.12VRR385 pKa = 11.84KK386 pKa = 8.28PQTPIEE392 pKa = 4.38ALLQQLVTASKK403 pKa = 10.81AQDD406 pKa = 3.36QEE408 pKa = 4.22HH409 pKa = 6.03TLFRR413 pKa = 11.84PKK415 pKa = 10.84GLLXLPAPEE424 pKa = 5.14ASMLNNPSTTFVPSAFSGFRR444 pKa = 11.84PIPPHH449 pKa = 5.2DD450 pKa = 4.02HH451 pKa = 6.78KK452 pKa = 11.27EE453 pKa = 4.06LVLEE457 pKa = 4.54EE458 pKa = 4.37PQPAFPPPPPPPPPPPPPPATMPVHH483 pKa = 4.93QRR485 pKa = 11.84KK486 pKa = 6.95TEE488 pKa = 4.12SVPFLTPPSSPIILGGPNNLLGRR511 pKa = 11.84TQAFLNEE518 pKa = 3.8IQQGKK523 pKa = 9.21KK524 pKa = 9.78LRR526 pKa = 11.84AAQDD530 pKa = 3.0RR531 pKa = 11.84MLAPLPVTKK540 pKa = 10.25HH541 pKa = 5.56GSPVADD547 pKa = 4.17FHH549 pKa = 7.26SQLADD554 pKa = 3.47VLKK557 pKa = 10.04TGPKK561 pKa = 9.14LRR563 pKa = 11.84KK564 pKa = 8.85VQPKK568 pKa = 8.51HH569 pKa = 4.16TVKK572 pKa = 10.81EE573 pKa = 4.15EE574 pKa = 4.05TMPTNPGDD582 pKa = 3.83RR583 pKa = 11.84IFNSLITKK591 pKa = 9.6IRR593 pKa = 11.84TRR595 pKa = 11.84VSPEE599 pKa = 3.48EE600 pKa = 4.16GLDD603 pKa = 3.5EE604 pKa = 5.65DD605 pKa = 4.14SWW607 pKa = 4.45
MM1 pKa = 7.77AKK3 pKa = 9.01TRR5 pKa = 11.84LLPSKK10 pKa = 10.64KK11 pKa = 9.44KK12 pKa = 10.52GKK14 pKa = 9.69KK15 pKa = 7.4HH16 pKa = 5.39TPNKK20 pKa = 9.94VKK22 pKa = 10.25IKK24 pKa = 10.38KK25 pKa = 8.79EE26 pKa = 3.98PKK28 pKa = 9.99SGITALRR35 pKa = 11.84SYY37 pKa = 11.16KK38 pKa = 10.52HH39 pKa = 5.53FTRR42 pKa = 11.84RR43 pKa = 11.84WNKK46 pKa = 10.25KK47 pKa = 5.85NTQEE51 pKa = 4.29KK52 pKa = 9.9SSPVQTLVKK61 pKa = 10.46VEE63 pKa = 4.34NNIQSSHH70 pKa = 5.73TGGKK74 pKa = 8.78QRR76 pKa = 11.84STMPKK81 pKa = 9.49LRR83 pKa = 11.84AKK85 pKa = 10.21PVVKK89 pKa = 10.26KK90 pKa = 8.89RR91 pKa = 11.84TRR93 pKa = 11.84GSVRR97 pKa = 11.84RR98 pKa = 11.84TPYY101 pKa = 9.43TKK103 pKa = 10.02PNHH106 pKa = 5.82SQASKK111 pKa = 10.5YY112 pKa = 10.58NSTRR116 pKa = 11.84ITKK119 pKa = 7.54STRR122 pKa = 11.84KK123 pKa = 8.3NTTTAPIITVSPNMTFNPQNSNVSSPRR150 pKa = 11.84VSNALVPYY158 pKa = 9.73NHH160 pKa = 6.32NQAQQGLRR168 pKa = 11.84RR169 pKa = 11.84MVEE172 pKa = 4.2KK173 pKa = 10.56EE174 pKa = 3.77SSAPGLASRR183 pKa = 11.84VLRR186 pKa = 11.84RR187 pKa = 11.84MINVALPTLGSAALASMTGIPSFLTNAAVNAVSSAITSDD226 pKa = 3.39EE227 pKa = 4.09DD228 pKa = 3.6VGPVTPTIPIYY239 pKa = 10.76HH240 pKa = 7.49EE241 pKa = 4.56STDD244 pKa = 3.59SAQPALRR251 pKa = 11.84YY252 pKa = 9.72QPIPQYY258 pKa = 11.11SGGPKK263 pKa = 9.72PDD265 pKa = 4.57DD266 pKa = 4.86DD267 pKa = 5.04SPGLLSTAQNLFEE280 pKa = 5.21NMLAYY285 pKa = 10.59VPTLTTLAGAGHH297 pKa = 6.93KK298 pKa = 10.04VYY300 pKa = 10.62QAMRR304 pKa = 11.84GGVAPYY310 pKa = 10.62VSGTHH315 pKa = 6.6TYY317 pKa = 9.2TPQASYY323 pKa = 10.42IQDD326 pKa = 3.33RR327 pKa = 11.84YY328 pKa = 10.3YY329 pKa = 10.92RR330 pKa = 11.84PAPYY334 pKa = 9.62ISSQQYY340 pKa = 9.78RR341 pKa = 11.84GGSGYY346 pKa = 9.96IPSAACRR353 pKa = 11.84VSVHH357 pKa = 5.99NPRR360 pKa = 11.84QYY362 pKa = 11.48ALRR365 pKa = 11.84GTKK368 pKa = 8.13HH369 pKa = 5.6TYY371 pKa = 10.04RR372 pKa = 11.84NRR374 pKa = 11.84AVHH377 pKa = 6.39NSGKK381 pKa = 10.46YY382 pKa = 7.77YY383 pKa = 10.12VRR385 pKa = 11.84KK386 pKa = 8.28PQTPIEE392 pKa = 4.38ALLQQLVTASKK403 pKa = 10.81AQDD406 pKa = 3.36QEE408 pKa = 4.22HH409 pKa = 6.03TLFRR413 pKa = 11.84PKK415 pKa = 10.84GLLXLPAPEE424 pKa = 5.14ASMLNNPSTTFVPSAFSGFRR444 pKa = 11.84PIPPHH449 pKa = 5.2DD450 pKa = 4.02HH451 pKa = 6.78KK452 pKa = 11.27EE453 pKa = 4.06LVLEE457 pKa = 4.54EE458 pKa = 4.37PQPAFPPPPPPPPPPPPPPATMPVHH483 pKa = 4.93QRR485 pKa = 11.84KK486 pKa = 6.95TEE488 pKa = 4.12SVPFLTPPSSPIILGGPNNLLGRR511 pKa = 11.84TQAFLNEE518 pKa = 3.8IQQGKK523 pKa = 9.21KK524 pKa = 9.78LRR526 pKa = 11.84AAQDD530 pKa = 3.0RR531 pKa = 11.84MLAPLPVTKK540 pKa = 10.25HH541 pKa = 5.56GSPVADD547 pKa = 4.17FHH549 pKa = 7.26SQLADD554 pKa = 3.47VLKK557 pKa = 10.04TGPKK561 pKa = 9.14LRR563 pKa = 11.84KK564 pKa = 8.85VQPKK568 pKa = 8.51HH569 pKa = 4.16TVKK572 pKa = 10.81EE573 pKa = 4.15EE574 pKa = 4.05TMPTNPGDD582 pKa = 3.83RR583 pKa = 11.84IFNSLITKK591 pKa = 9.6IRR593 pKa = 11.84TRR595 pKa = 11.84VSPEE599 pKa = 3.48EE600 pKa = 4.16GLDD603 pKa = 3.5EE604 pKa = 5.65DD605 pKa = 4.14SWW607 pKa = 4.45
Molecular weight: 66.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5200 |
172 |
1101 |
520.0 |
57.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.904 ± 0.398 | 2.692 ± 0.43 |
4.423 ± 0.397 | 5.942 ± 0.594 |
3.615 ± 0.466 | 5.692 ± 0.457 |
3.481 ± 0.221 | 4.019 ± 0.357 |
4.654 ± 0.51 | 8.115 ± 0.306 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.5 ± 0.312 | 4.154 ± 0.214 |
6.827 ± 0.834 | 4.635 ± 0.318 |
6.308 ± 0.5 | 8.154 ± 0.478 |
7.077 ± 0.535 | 7.269 ± 0.309 |
0.75 ± 0.168 | 2.769 ± 0.267 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
