
Torque teno virus 29
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.65
Get precalculated fractions of proteins
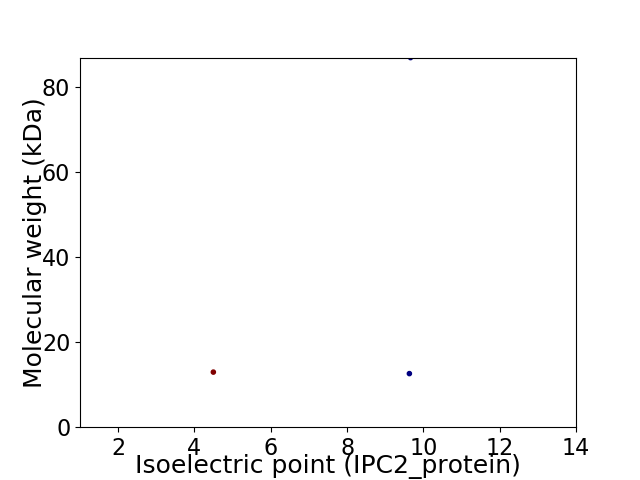
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9JG77|Q9JG77_9VIRU ORF2 ORF1 ORF3 genes isolate: TTVyon-KC009 OS=Torque teno virus 29 OX=687368 PE=4 SV=1
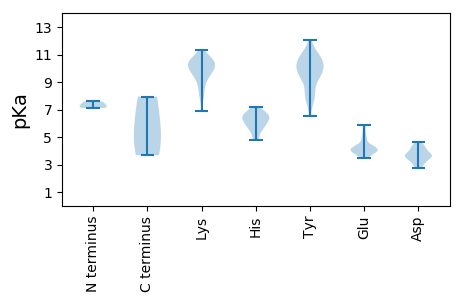
MM1 pKa = 7.59PWRR4 pKa = 11.84PPGHH8 pKa = 5.95NVPGRR13 pKa = 11.84EE14 pKa = 4.15NQWFAAVFHH23 pKa = 6.52SHH25 pKa = 7.17ASWCGCGDD33 pKa = 3.71FVGHH37 pKa = 7.16LNSIAPRR44 pKa = 11.84FSNAGPPRR52 pKa = 11.84PPPGLDD58 pKa = 2.87QHH60 pKa = 6.82NPEE63 pKa = 4.84GPAGPGGPPAILPALPAPADD83 pKa = 3.67PEE85 pKa = 4.24PPPRR89 pKa = 11.84RR90 pKa = 11.84GGGADD95 pKa = 3.14GGAAGGLAIADD106 pKa = 3.64APGGYY111 pKa = 10.48GEE113 pKa = 5.89DD114 pKa = 4.38DD115 pKa = 3.98LDD117 pKa = 3.94EE118 pKa = 4.96LFAAAAEE125 pKa = 4.25DD126 pKa = 4.55DD127 pKa = 3.84MM128 pKa = 7.94
MM1 pKa = 7.59PWRR4 pKa = 11.84PPGHH8 pKa = 5.95NVPGRR13 pKa = 11.84EE14 pKa = 4.15NQWFAAVFHH23 pKa = 6.52SHH25 pKa = 7.17ASWCGCGDD33 pKa = 3.71FVGHH37 pKa = 7.16LNSIAPRR44 pKa = 11.84FSNAGPPRR52 pKa = 11.84PPPGLDD58 pKa = 2.87QHH60 pKa = 6.82NPEE63 pKa = 4.84GPAGPGGPPAILPALPAPADD83 pKa = 3.67PEE85 pKa = 4.24PPPRR89 pKa = 11.84RR90 pKa = 11.84GGGADD95 pKa = 3.14GGAAGGLAIADD106 pKa = 3.64APGGYY111 pKa = 10.48GEE113 pKa = 5.89DD114 pKa = 4.38DD115 pKa = 3.98LDD117 pKa = 3.94EE118 pKa = 4.96LFAAAAEE125 pKa = 4.25DD126 pKa = 4.55DD127 pKa = 3.84MM128 pKa = 7.94
Molecular weight: 12.95 kDa
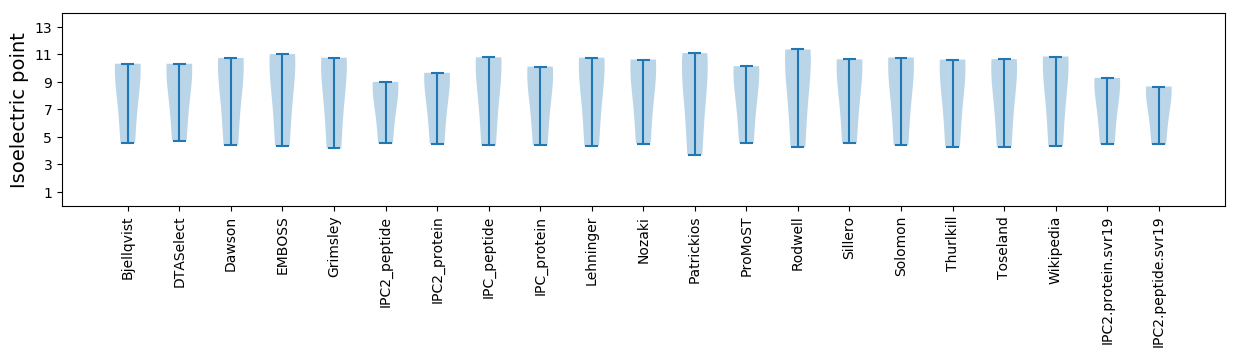
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9JG77|Q9JG77_9VIRU ORF2 ORF1 ORF3 genes isolate: TTVyon-KC009 OS=Torque teno virus 29 OX=687368 PE=4 SV=1
MM1 pKa = 7.11AWWWGRR7 pKa = 11.84WRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84PRR15 pKa = 11.84YY16 pKa = 9.39RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TWRR22 pKa = 11.84VRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84RR30 pKa = 11.84TFRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84YY40 pKa = 6.52VSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 7.93YY50 pKa = 9.88RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84LRR55 pKa = 11.84RR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84GRR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 9.26RR65 pKa = 11.84HH66 pKa = 4.96RR67 pKa = 11.84QTLILRR73 pKa = 11.84QWQPDD78 pKa = 3.71VVRR81 pKa = 11.84HH82 pKa = 5.17CKK84 pKa = 8.0ITGWMPLIICGSGSTQNNFITHH106 pKa = 6.46MDD108 pKa = 4.1DD109 pKa = 4.15FPPMGYY115 pKa = 9.9SYY117 pKa = 11.47GGNFTNLTFSLEE129 pKa = 4.26GIYY132 pKa = 10.33EE133 pKa = 3.89QFLYY137 pKa = 10.43HH138 pKa = 6.71RR139 pKa = 11.84NRR141 pKa = 11.84WSRR144 pKa = 11.84SNHH147 pKa = 5.98DD148 pKa = 4.26LDD150 pKa = 3.61LARR153 pKa = 11.84YY154 pKa = 9.37KK155 pKa = 10.44GTTLKK160 pKa = 10.69LYY162 pKa = 10.21RR163 pKa = 11.84HH164 pKa = 6.14HH165 pKa = 6.48TLDD168 pKa = 4.29YY169 pKa = 9.38IVSYY173 pKa = 11.06NRR175 pKa = 11.84TGPFQISDD183 pKa = 3.38MTYY186 pKa = 10.9LSTHH190 pKa = 6.66PALMLLQKK198 pKa = 10.46HH199 pKa = 6.11RR200 pKa = 11.84IVVPSLLTKK209 pKa = 10.51PKK211 pKa = 9.45GKK213 pKa = 10.17RR214 pKa = 11.84SIKK217 pKa = 10.33VRR219 pKa = 11.84IKK221 pKa = 10.4PPKK224 pKa = 10.15LMLNKK229 pKa = 9.61WYY231 pKa = 7.82FTKK234 pKa = 10.49DD235 pKa = 2.96ICSMGLFQLQATACTLYY252 pKa = 10.93NPWLRR257 pKa = 11.84DD258 pKa = 3.56TTKK261 pKa = 10.86SPVIASRR268 pKa = 11.84VLKK271 pKa = 10.85NSIYY275 pKa = 10.42TNLSNLPEE283 pKa = 4.45HH284 pKa = 6.44NGARR288 pKa = 11.84EE289 pKa = 4.02SIRR292 pKa = 11.84NKK294 pKa = 10.09LHH296 pKa = 6.6PEE298 pKa = 3.94SLTGSVPNQKK308 pKa = 9.38GWEE311 pKa = 3.95YY312 pKa = 11.13SYY314 pKa = 10.75TKK316 pKa = 10.83LMAPIYY322 pKa = 9.49YY323 pKa = 9.47QANRR327 pKa = 11.84NSTYY331 pKa = 11.01NWLNYY336 pKa = 7.47QQNYY340 pKa = 7.81SQTYY344 pKa = 7.69QAFKK348 pKa = 11.13QKK350 pKa = 9.88MNNNLTLIQQEE361 pKa = 4.35YY362 pKa = 8.38MYY364 pKa = 9.71HH365 pKa = 5.73YY366 pKa = 9.16PNSVTTDD373 pKa = 3.55IIGKK377 pKa = 8.24NTLTHH382 pKa = 6.78DD383 pKa = 3.08WGIYY387 pKa = 8.32SPYY390 pKa = 9.45WLTPTRR396 pKa = 11.84ISLDD400 pKa = 2.74WGTPWTYY407 pKa = 10.82VRR409 pKa = 11.84YY410 pKa = 10.13NPLADD415 pKa = 3.6KK416 pKa = 11.29GIGNAVYY423 pKa = 9.95AQWCSEE429 pKa = 3.93QTSNLDD435 pKa = 3.57TKK437 pKa = 10.78KK438 pKa = 10.54SKK440 pKa = 10.87CIMKK444 pKa = 9.92DD445 pKa = 3.23LPLWCIFYY453 pKa = 10.56GYY455 pKa = 10.31VDD457 pKa = 4.66WIIKK461 pKa = 7.95STGVSSAVTDD471 pKa = 3.37MRR473 pKa = 11.84VAIISPYY480 pKa = 9.39TEE482 pKa = 3.94PALIGSSPEE491 pKa = 3.51VGYY494 pKa = 10.64IPVSDD499 pKa = 4.04TFCNGDD505 pKa = 3.66MPFLAPYY512 pKa = 9.89IPVGWWIKK520 pKa = 7.84WYY522 pKa = 10.19PMIAHH527 pKa = 5.6QKK529 pKa = 9.04EE530 pKa = 4.23VFEE533 pKa = 5.65AIVNCGPFVPRR544 pKa = 11.84DD545 pKa = 3.32QTTPSWEE552 pKa = 3.47ITMGYY557 pKa = 9.9KK558 pKa = 9.23MDD560 pKa = 4.01WLWGGSPLPSQAIDD574 pKa = 4.3DD575 pKa = 4.41PCQKK579 pKa = 8.79PTHH582 pKa = 6.25EE583 pKa = 4.97LPDD586 pKa = 4.35PDD588 pKa = 3.36RR589 pKa = 11.84HH590 pKa = 5.65PRR592 pKa = 11.84MLQVSDD598 pKa = 3.74PTKK601 pKa = 10.63LGPKK605 pKa = 8.7TVFHH609 pKa = 6.29KK610 pKa = 10.2WDD612 pKa = 3.07WRR614 pKa = 11.84RR615 pKa = 11.84GMLSKK620 pKa = 10.78RR621 pKa = 11.84SIKK624 pKa = 10.11RR625 pKa = 11.84VQEE628 pKa = 4.12DD629 pKa = 3.76STDD632 pKa = 3.59DD633 pKa = 3.82EE634 pKa = 5.04YY635 pKa = 12.07VAGPLPRR642 pKa = 11.84KK643 pKa = 9.56RR644 pKa = 11.84NKK646 pKa = 9.93FDD648 pKa = 2.98TRR650 pKa = 11.84AQGLQSPEE658 pKa = 3.92KK659 pKa = 10.44EE660 pKa = 4.08NYY662 pKa = 8.52TLLQALQDD670 pKa = 3.91SGQEE674 pKa = 3.98SSSEE678 pKa = 4.05NQEE681 pKa = 3.59QAPQEE686 pKa = 4.12KK687 pKa = 9.78EE688 pKa = 4.18GQKK691 pKa = 9.9EE692 pKa = 3.78ALMEE696 pKa = 3.76QLQLQKK702 pKa = 10.39QHH704 pKa = 5.57QRR706 pKa = 11.84VLKK709 pKa = 10.59RR710 pKa = 11.84GLKK713 pKa = 10.3LLLGDD718 pKa = 3.49VLRR721 pKa = 11.84LRR723 pKa = 11.84RR724 pKa = 11.84GVHH727 pKa = 5.93WDD729 pKa = 3.37PLLSS733 pKa = 3.68
MM1 pKa = 7.11AWWWGRR7 pKa = 11.84WRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84PRR15 pKa = 11.84YY16 pKa = 9.39RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TWRR22 pKa = 11.84VRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84PRR29 pKa = 11.84RR30 pKa = 11.84TFRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84YY40 pKa = 6.52VSRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 7.93YY50 pKa = 9.88RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84LRR55 pKa = 11.84RR56 pKa = 11.84GRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84GRR62 pKa = 11.84RR63 pKa = 11.84KK64 pKa = 9.26RR65 pKa = 11.84HH66 pKa = 4.96RR67 pKa = 11.84QTLILRR73 pKa = 11.84QWQPDD78 pKa = 3.71VVRR81 pKa = 11.84HH82 pKa = 5.17CKK84 pKa = 8.0ITGWMPLIICGSGSTQNNFITHH106 pKa = 6.46MDD108 pKa = 4.1DD109 pKa = 4.15FPPMGYY115 pKa = 9.9SYY117 pKa = 11.47GGNFTNLTFSLEE129 pKa = 4.26GIYY132 pKa = 10.33EE133 pKa = 3.89QFLYY137 pKa = 10.43HH138 pKa = 6.71RR139 pKa = 11.84NRR141 pKa = 11.84WSRR144 pKa = 11.84SNHH147 pKa = 5.98DD148 pKa = 4.26LDD150 pKa = 3.61LARR153 pKa = 11.84YY154 pKa = 9.37KK155 pKa = 10.44GTTLKK160 pKa = 10.69LYY162 pKa = 10.21RR163 pKa = 11.84HH164 pKa = 6.14HH165 pKa = 6.48TLDD168 pKa = 4.29YY169 pKa = 9.38IVSYY173 pKa = 11.06NRR175 pKa = 11.84TGPFQISDD183 pKa = 3.38MTYY186 pKa = 10.9LSTHH190 pKa = 6.66PALMLLQKK198 pKa = 10.46HH199 pKa = 6.11RR200 pKa = 11.84IVVPSLLTKK209 pKa = 10.51PKK211 pKa = 9.45GKK213 pKa = 10.17RR214 pKa = 11.84SIKK217 pKa = 10.33VRR219 pKa = 11.84IKK221 pKa = 10.4PPKK224 pKa = 10.15LMLNKK229 pKa = 9.61WYY231 pKa = 7.82FTKK234 pKa = 10.49DD235 pKa = 2.96ICSMGLFQLQATACTLYY252 pKa = 10.93NPWLRR257 pKa = 11.84DD258 pKa = 3.56TTKK261 pKa = 10.86SPVIASRR268 pKa = 11.84VLKK271 pKa = 10.85NSIYY275 pKa = 10.42TNLSNLPEE283 pKa = 4.45HH284 pKa = 6.44NGARR288 pKa = 11.84EE289 pKa = 4.02SIRR292 pKa = 11.84NKK294 pKa = 10.09LHH296 pKa = 6.6PEE298 pKa = 3.94SLTGSVPNQKK308 pKa = 9.38GWEE311 pKa = 3.95YY312 pKa = 11.13SYY314 pKa = 10.75TKK316 pKa = 10.83LMAPIYY322 pKa = 9.49YY323 pKa = 9.47QANRR327 pKa = 11.84NSTYY331 pKa = 11.01NWLNYY336 pKa = 7.47QQNYY340 pKa = 7.81SQTYY344 pKa = 7.69QAFKK348 pKa = 11.13QKK350 pKa = 9.88MNNNLTLIQQEE361 pKa = 4.35YY362 pKa = 8.38MYY364 pKa = 9.71HH365 pKa = 5.73YY366 pKa = 9.16PNSVTTDD373 pKa = 3.55IIGKK377 pKa = 8.24NTLTHH382 pKa = 6.78DD383 pKa = 3.08WGIYY387 pKa = 8.32SPYY390 pKa = 9.45WLTPTRR396 pKa = 11.84ISLDD400 pKa = 2.74WGTPWTYY407 pKa = 10.82VRR409 pKa = 11.84YY410 pKa = 10.13NPLADD415 pKa = 3.6KK416 pKa = 11.29GIGNAVYY423 pKa = 9.95AQWCSEE429 pKa = 3.93QTSNLDD435 pKa = 3.57TKK437 pKa = 10.78KK438 pKa = 10.54SKK440 pKa = 10.87CIMKK444 pKa = 9.92DD445 pKa = 3.23LPLWCIFYY453 pKa = 10.56GYY455 pKa = 10.31VDD457 pKa = 4.66WIIKK461 pKa = 7.95STGVSSAVTDD471 pKa = 3.37MRR473 pKa = 11.84VAIISPYY480 pKa = 9.39TEE482 pKa = 3.94PALIGSSPEE491 pKa = 3.51VGYY494 pKa = 10.64IPVSDD499 pKa = 4.04TFCNGDD505 pKa = 3.66MPFLAPYY512 pKa = 9.89IPVGWWIKK520 pKa = 7.84WYY522 pKa = 10.19PMIAHH527 pKa = 5.6QKK529 pKa = 9.04EE530 pKa = 4.23VFEE533 pKa = 5.65AIVNCGPFVPRR544 pKa = 11.84DD545 pKa = 3.32QTTPSWEE552 pKa = 3.47ITMGYY557 pKa = 9.9KK558 pKa = 9.23MDD560 pKa = 4.01WLWGGSPLPSQAIDD574 pKa = 4.3DD575 pKa = 4.41PCQKK579 pKa = 8.79PTHH582 pKa = 6.25EE583 pKa = 4.97LPDD586 pKa = 4.35PDD588 pKa = 3.36RR589 pKa = 11.84HH590 pKa = 5.65PRR592 pKa = 11.84MLQVSDD598 pKa = 3.74PTKK601 pKa = 10.63LGPKK605 pKa = 8.7TVFHH609 pKa = 6.29KK610 pKa = 10.2WDD612 pKa = 3.07WRR614 pKa = 11.84RR615 pKa = 11.84GMLSKK620 pKa = 10.78RR621 pKa = 11.84SIKK624 pKa = 10.11RR625 pKa = 11.84VQEE628 pKa = 4.12DD629 pKa = 3.76STDD632 pKa = 3.59DD633 pKa = 3.82EE634 pKa = 5.04YY635 pKa = 12.07VAGPLPRR642 pKa = 11.84KK643 pKa = 9.56RR644 pKa = 11.84NKK646 pKa = 9.93FDD648 pKa = 2.98TRR650 pKa = 11.84AQGLQSPEE658 pKa = 3.92KK659 pKa = 10.44EE660 pKa = 4.08NYY662 pKa = 8.52TLLQALQDD670 pKa = 3.91SGQEE674 pKa = 3.98SSSEE678 pKa = 4.05NQEE681 pKa = 3.59QAPQEE686 pKa = 4.12KK687 pKa = 9.78EE688 pKa = 4.18GQKK691 pKa = 9.9EE692 pKa = 3.78ALMEE696 pKa = 3.76QLQLQKK702 pKa = 10.39QHH704 pKa = 5.57QRR706 pKa = 11.84VLKK709 pKa = 10.59RR710 pKa = 11.84GLKK713 pKa = 10.3LLLGDD718 pKa = 3.49VLRR721 pKa = 11.84LRR723 pKa = 11.84RR724 pKa = 11.84GVHH727 pKa = 5.93WDD729 pKa = 3.37PLLSS733 pKa = 3.68
Molecular weight: 86.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
975 |
114 |
733 |
325.0 |
37.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.231 ± 2.984 | 1.333 ± 0.103 |
4.615 ± 0.912 | 4.0 ± 0.95 |
2.462 ± 0.345 | 6.974 ± 2.378 |
2.667 ± 0.298 | 4.205 ± 1.288 |
6.154 ± 1.867 | 7.795 ± 1.643 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.564 ± 0.277 | 4.41 ± 0.148 |
8.41 ± 2.796 | 4.615 ± 1.262 |
9.026 ± 1.617 | 8.0 ± 3.239 |
5.949 ± 1.473 | 3.59 ± 0.988 |
3.385 ± 0.889 | 4.615 ± 1.474 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
