
Calocera viscosa (strain TUFC12733)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Dacrymycetes; Dacrymycetales; Dacrymycetaceae; Calocera; Calocera viscosa
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
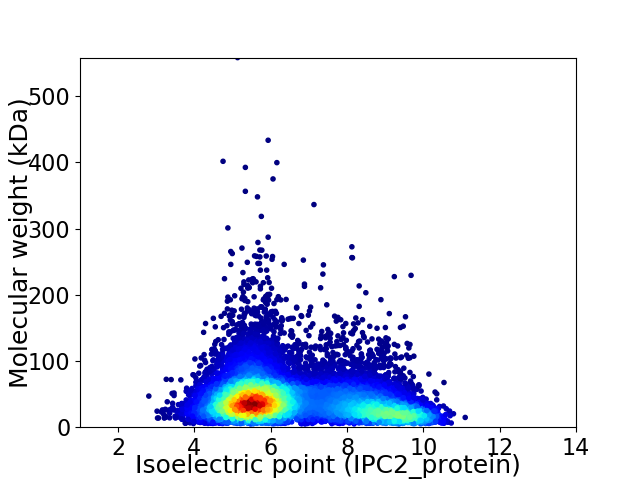
Virtual 2D-PAGE plot for 12323 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
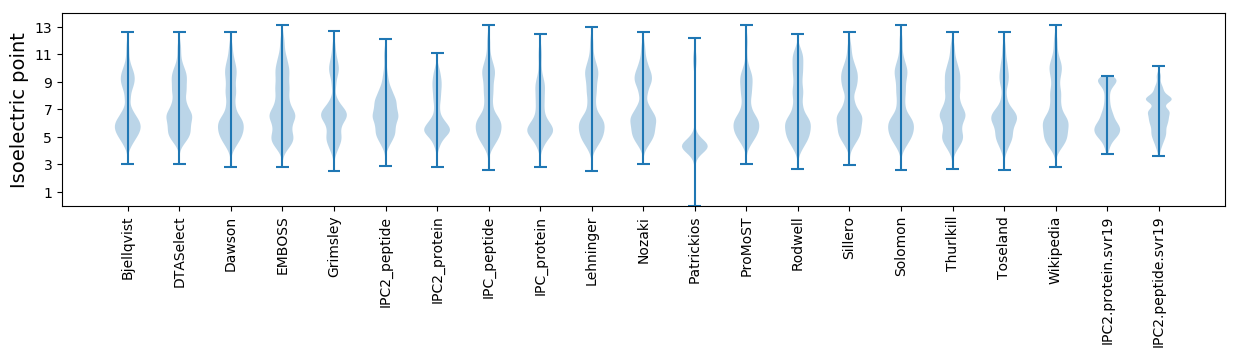
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A167HV73|A0A167HV73_CALVF Uncharacterized protein (Fragment) OS=Calocera viscosa (strain TUFC12733) OX=1330018 GN=CALVIDRAFT_463058 PE=4 SV=1
MM1 pKa = 7.01QRR3 pKa = 11.84LSALVLALLSASQLAHH19 pKa = 6.96ALPLRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 4.28VPQEE30 pKa = 3.98HH31 pKa = 6.06SHH33 pKa = 5.98QPFLTSVQTSLQLNNPDD50 pKa = 4.75GITDD54 pKa = 3.75SVFGLLGDD62 pKa = 4.22AAAAAGAGKK71 pKa = 8.25ITDD74 pKa = 3.94MDD76 pKa = 4.44CLQQATADD84 pKa = 3.63QAFTNAKK91 pKa = 9.95AAGDD95 pKa = 3.64VQGQVDD101 pKa = 3.91ALIYY105 pKa = 10.19RR106 pKa = 11.84ALEE109 pKa = 4.22RR110 pKa = 11.84NTGAIGLASAACTSLTAVNPEE131 pKa = 3.4IAAIAQHH138 pKa = 5.94QDD140 pKa = 3.26PASSNAAAVNKK151 pKa = 10.55AIVLTLAQQIAAVGGNPLDD170 pKa = 4.76AIKK173 pKa = 10.75SGTFAPGTIGDD184 pKa = 3.82PTAAGNTCDD193 pKa = 5.38DD194 pKa = 4.24ANDD197 pKa = 3.79ANGCIFTQNLLVPDD211 pKa = 4.64ASDD214 pKa = 4.37DD215 pKa = 4.35EE216 pKa = 4.96IDD218 pKa = 3.64AAVAGISSSSGNSTTTDD235 pKa = 2.9SSTGSTDD242 pKa = 3.08TSSTDD247 pKa = 3.19SDD249 pKa = 4.38SDD251 pKa = 3.86TATCPPPSTVTVTVTSTAAAEE272 pKa = 4.19TATGSSSAAAATSTAISTASAGTGSSTASSNGTNLQTFTGALGGILPPAVVAGGEE327 pKa = 4.46GFITDD332 pKa = 3.67NGSQFVNLAAALGRR346 pKa = 11.84SCDD349 pKa = 3.71VQHH352 pKa = 6.16NQCADD357 pKa = 3.2QANSQNQSPFSVGDD371 pKa = 3.98CDD373 pKa = 4.8TQDD376 pKa = 4.05TEE378 pKa = 4.69CKK380 pKa = 10.19AAGTGGSTTGTSAASSSTASTTTTQAAAVGATSGSISTVATSGTNLQTFTGALGGILPPPVVAGGEE446 pKa = 4.27GFITDD451 pKa = 3.67NGSQFVNLAAALGRR465 pKa = 11.84SCDD468 pKa = 3.67MQHH471 pKa = 5.92NQCADD476 pKa = 3.61LANSQHH482 pKa = 5.54QFSVSDD488 pKa = 4.24CDD490 pKa = 4.52QQDD493 pKa = 4.2TEE495 pKa = 4.78CKK497 pKa = 10.09AAA499 pKa = 4.27
MM1 pKa = 7.01QRR3 pKa = 11.84LSALVLALLSASQLAHH19 pKa = 6.96ALPLRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 4.28VPQEE30 pKa = 3.98HH31 pKa = 6.06SHH33 pKa = 5.98QPFLTSVQTSLQLNNPDD50 pKa = 4.75GITDD54 pKa = 3.75SVFGLLGDD62 pKa = 4.22AAAAAGAGKK71 pKa = 8.25ITDD74 pKa = 3.94MDD76 pKa = 4.44CLQQATADD84 pKa = 3.63QAFTNAKK91 pKa = 9.95AAGDD95 pKa = 3.64VQGQVDD101 pKa = 3.91ALIYY105 pKa = 10.19RR106 pKa = 11.84ALEE109 pKa = 4.22RR110 pKa = 11.84NTGAIGLASAACTSLTAVNPEE131 pKa = 3.4IAAIAQHH138 pKa = 5.94QDD140 pKa = 3.26PASSNAAAVNKK151 pKa = 10.55AIVLTLAQQIAAVGGNPLDD170 pKa = 4.76AIKK173 pKa = 10.75SGTFAPGTIGDD184 pKa = 3.82PTAAGNTCDD193 pKa = 5.38DD194 pKa = 4.24ANDD197 pKa = 3.79ANGCIFTQNLLVPDD211 pKa = 4.64ASDD214 pKa = 4.37DD215 pKa = 4.35EE216 pKa = 4.96IDD218 pKa = 3.64AAVAGISSSSGNSTTTDD235 pKa = 2.9SSTGSTDD242 pKa = 3.08TSSTDD247 pKa = 3.19SDD249 pKa = 4.38SDD251 pKa = 3.86TATCPPPSTVTVTVTSTAAAEE272 pKa = 4.19TATGSSSAAAATSTAISTASAGTGSSTASSNGTNLQTFTGALGGILPPAVVAGGEE327 pKa = 4.46GFITDD332 pKa = 3.67NGSQFVNLAAALGRR346 pKa = 11.84SCDD349 pKa = 3.71VQHH352 pKa = 6.16NQCADD357 pKa = 3.2QANSQNQSPFSVGDD371 pKa = 3.98CDD373 pKa = 4.8TQDD376 pKa = 4.05TEE378 pKa = 4.69CKK380 pKa = 10.19AAGTGGSTTGTSAASSSTASTTTTQAAAVGATSGSISTVATSGTNLQTFTGALGGILPPPVVAGGEE446 pKa = 4.27GFITDD451 pKa = 3.67NGSQFVNLAAALGRR465 pKa = 11.84SCDD468 pKa = 3.67MQHH471 pKa = 5.92NQCADD476 pKa = 3.61LANSQHH482 pKa = 5.54QFSVSDD488 pKa = 4.24CDD490 pKa = 4.52QQDD493 pKa = 4.2TEE495 pKa = 4.78CKK497 pKa = 10.09AAA499 pKa = 4.27
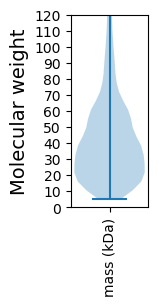
Molecular weight: 49.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A167LED9|A0A167LED9_CALVF Uncharacterized protein OS=Calocera viscosa (strain TUFC12733) OX=1330018 GN=CALVIDRAFT_537994 PE=4 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84HH4 pKa = 6.71LIRR7 pKa = 11.84RR8 pKa = 11.84AITAAARR15 pKa = 11.84TATRR19 pKa = 11.84PAAPTTTAVSAVALVRR35 pKa = 11.84PSSSRR40 pKa = 11.84IGTSTLPSPFRR51 pKa = 11.84TLTPFSPRR59 pKa = 11.84PSALTTLSPVRR70 pKa = 11.84PSIALSPLTAALPLLQARR88 pKa = 11.84FLGNEE93 pKa = 3.95YY94 pKa = 10.37QPSQRR99 pKa = 11.84KK100 pKa = 8.82RR101 pKa = 11.84KK102 pKa = 9.39RR103 pKa = 11.84KK104 pKa = 9.34FGFLQRR110 pKa = 11.84LRR112 pKa = 11.84SKK114 pKa = 10.83NGRR117 pKa = 11.84KK118 pKa = 7.28TLAARR123 pKa = 11.84RR124 pKa = 11.84WKK126 pKa = 9.4GRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 9.93LSHH133 pKa = 7.01
MM1 pKa = 7.54PRR3 pKa = 11.84HH4 pKa = 6.71LIRR7 pKa = 11.84RR8 pKa = 11.84AITAAARR15 pKa = 11.84TATRR19 pKa = 11.84PAAPTTTAVSAVALVRR35 pKa = 11.84PSSSRR40 pKa = 11.84IGTSTLPSPFRR51 pKa = 11.84TLTPFSPRR59 pKa = 11.84PSALTTLSPVRR70 pKa = 11.84PSIALSPLTAALPLLQARR88 pKa = 11.84FLGNEE93 pKa = 3.95YY94 pKa = 10.37QPSQRR99 pKa = 11.84KK100 pKa = 8.82RR101 pKa = 11.84KK102 pKa = 9.39RR103 pKa = 11.84KK104 pKa = 9.34FGFLQRR110 pKa = 11.84LRR112 pKa = 11.84SKK114 pKa = 10.83NGRR117 pKa = 11.84KK118 pKa = 7.28TLAARR123 pKa = 11.84RR124 pKa = 11.84WKK126 pKa = 9.4GRR128 pKa = 11.84RR129 pKa = 11.84YY130 pKa = 9.93LSHH133 pKa = 7.01
Molecular weight: 14.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5084671 |
49 |
5013 |
412.6 |
45.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.386 ± 0.023 | 1.257 ± 0.009 |
5.323 ± 0.017 | 6.157 ± 0.025 |
3.483 ± 0.016 | 7.059 ± 0.02 |
2.426 ± 0.01 | 4.419 ± 0.015 |
4.133 ± 0.021 | 9.6 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.234 ± 0.009 | 2.999 ± 0.012 |
6.895 ± 0.03 | 3.798 ± 0.017 |
6.503 ± 0.021 | 7.886 ± 0.025 |
5.834 ± 0.013 | 6.391 ± 0.016 |
1.581 ± 0.009 | 2.634 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
