
Arachis hypogaea (Peanut)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade; dalbergioids sensu lato;
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
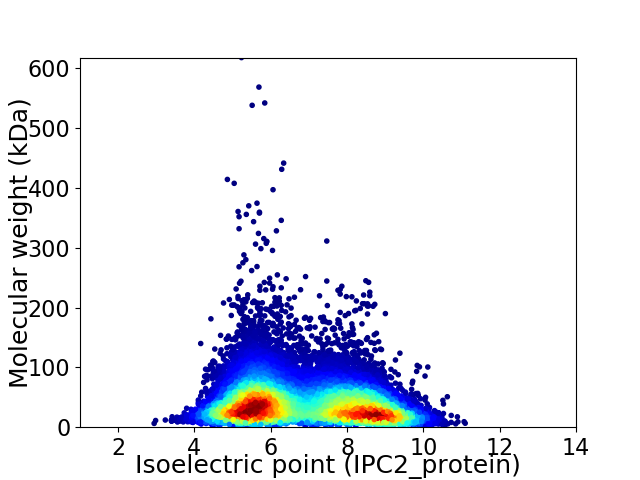
Virtual 2D-PAGE plot for 97596 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A445E6T8|A0A445E6T8_ARAHY Uncharacterized protein OS=Arachis hypogaea OX=3818 GN=Ahy_A02g005435 PE=4 SV=1
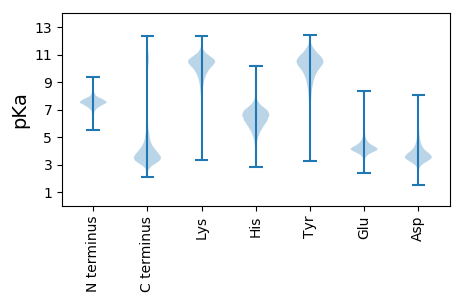
MM1 pKa = 7.77DD2 pKa = 3.54VKK4 pKa = 11.08GNNVTPDD11 pKa = 3.76FSSYY15 pKa = 11.15ILFEE19 pKa = 4.3ASGDD23 pKa = 3.89SEE25 pKa = 4.12EE26 pKa = 6.15ADD28 pKa = 3.75FNHH31 pKa = 6.24NKK33 pKa = 8.43MVCEE37 pKa = 4.36ISRR40 pKa = 11.84VGSYY44 pKa = 7.33VHH46 pKa = 6.96HH47 pKa = 6.22GHH49 pKa = 6.84EE50 pKa = 5.63DD51 pKa = 3.52YY52 pKa = 11.55DD53 pKa = 5.52DD54 pKa = 6.14DD55 pKa = 7.24DD56 pKa = 5.42DD57 pKa = 6.0ALSCSYY63 pKa = 10.96DD64 pKa = 3.59RR65 pKa = 11.84SDD67 pKa = 5.13YY68 pKa = 11.59DD69 pKa = 5.26DD70 pKa = 4.85DD71 pKa = 4.18ACNVAEE77 pKa = 4.81VNWDD81 pKa = 4.18DD82 pKa = 6.48DD83 pKa = 5.97DD84 pKa = 7.68DD85 pKa = 7.44DD86 pKa = 7.49DD87 pKa = 7.49DD88 pKa = 7.06DD89 pKa = 5.62GEE91 pKa = 4.48KK92 pKa = 10.44KK93 pKa = 10.58DD94 pKa = 4.64DD95 pKa = 3.73VFGTSYY101 pKa = 10.93CDD103 pKa = 3.56EE104 pKa = 4.45EE105 pKa = 6.15DD106 pKa = 3.67EE107 pKa = 4.75VLQEE111 pKa = 3.83QHH113 pKa = 5.93PTRR116 pKa = 11.84SFVSFASNQEE126 pKa = 4.17SLDD129 pKa = 3.6EE130 pKa = 4.11MEE132 pKa = 4.68KK133 pKa = 10.72NRR135 pKa = 11.84LFWEE139 pKa = 4.55ACLASS144 pKa = 3.65
MM1 pKa = 7.77DD2 pKa = 3.54VKK4 pKa = 11.08GNNVTPDD11 pKa = 3.76FSSYY15 pKa = 11.15ILFEE19 pKa = 4.3ASGDD23 pKa = 3.89SEE25 pKa = 4.12EE26 pKa = 6.15ADD28 pKa = 3.75FNHH31 pKa = 6.24NKK33 pKa = 8.43MVCEE37 pKa = 4.36ISRR40 pKa = 11.84VGSYY44 pKa = 7.33VHH46 pKa = 6.96HH47 pKa = 6.22GHH49 pKa = 6.84EE50 pKa = 5.63DD51 pKa = 3.52YY52 pKa = 11.55DD53 pKa = 5.52DD54 pKa = 6.14DD55 pKa = 7.24DD56 pKa = 5.42DD57 pKa = 6.0ALSCSYY63 pKa = 10.96DD64 pKa = 3.59RR65 pKa = 11.84SDD67 pKa = 5.13YY68 pKa = 11.59DD69 pKa = 5.26DD70 pKa = 4.85DD71 pKa = 4.18ACNVAEE77 pKa = 4.81VNWDD81 pKa = 4.18DD82 pKa = 6.48DD83 pKa = 5.97DD84 pKa = 7.68DD85 pKa = 7.44DD86 pKa = 7.49DD87 pKa = 7.49DD88 pKa = 7.06DD89 pKa = 5.62GEE91 pKa = 4.48KK92 pKa = 10.44KK93 pKa = 10.58DD94 pKa = 4.64DD95 pKa = 3.73VFGTSYY101 pKa = 10.93CDD103 pKa = 3.56EE104 pKa = 4.45EE105 pKa = 6.15DD106 pKa = 3.67EE107 pKa = 4.75VLQEE111 pKa = 3.83QHH113 pKa = 5.93PTRR116 pKa = 11.84SFVSFASNQEE126 pKa = 4.17SLDD129 pKa = 3.6EE130 pKa = 4.11MEE132 pKa = 4.68KK133 pKa = 10.72NRR135 pKa = 11.84LFWEE139 pKa = 4.55ACLASS144 pKa = 3.65
Molecular weight: 16.44 kDa
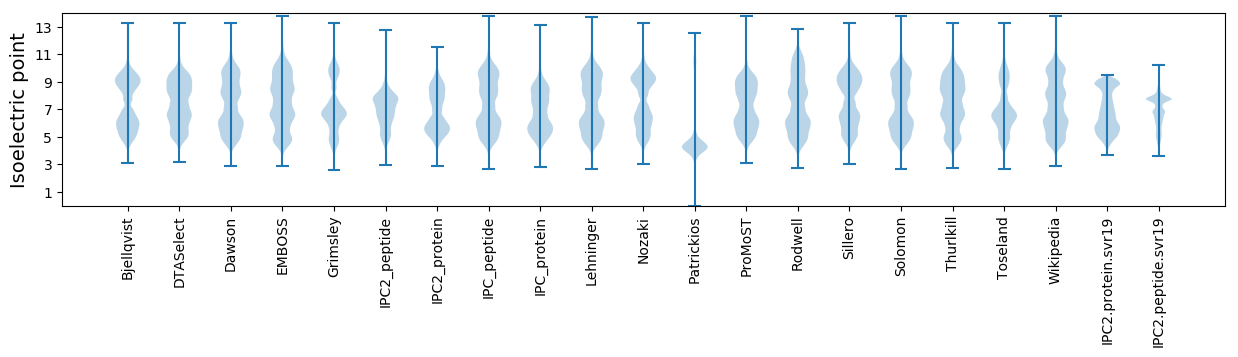
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A445CE00|A0A445CE00_ARAHY Isoform of A0A445CDQ1 Protein FAR1-RELATED SEQUENCE OS=Arachis hypogaea OX=3818 GN=Ahy_A07g035374 PE=3 SV=1
MM1 pKa = 7.53LVTLQRR7 pKa = 11.84RR8 pKa = 11.84QGALQRR14 pKa = 11.84MLGSLQRR21 pKa = 11.84KK22 pKa = 9.15LGALQRR28 pKa = 11.84WLGALQRR35 pKa = 11.84RR36 pKa = 11.84QVALQRR42 pKa = 11.84MLGSLQRR49 pKa = 11.84MLRR52 pKa = 11.84ALQRR56 pKa = 11.84LLGALQRR63 pKa = 11.84RR64 pKa = 11.84QVALQRR70 pKa = 11.84RR71 pKa = 11.84QGASQRR77 pKa = 11.84WLGVLQRR84 pKa = 11.84WLGALQRR91 pKa = 11.84RR92 pKa = 11.84QVALQRR98 pKa = 11.84WLRR101 pKa = 11.84AVQRR105 pKa = 11.84MLGALQRR112 pKa = 11.84WLGALQRR119 pKa = 11.84WLEE122 pKa = 3.99ALQRR126 pKa = 11.84RR127 pKa = 11.84QGALQRR133 pKa = 11.84RR134 pKa = 11.84QGALQRR140 pKa = 11.84WPVTLQRR147 pKa = 11.84KK148 pKa = 8.14QVDD151 pKa = 4.12LQRR154 pKa = 11.84MLGSLQRR161 pKa = 11.84KK162 pKa = 9.15LGALQRR168 pKa = 11.84WLGALQRR175 pKa = 11.84MLSALQRR182 pKa = 11.84WLVTLQRR189 pKa = 11.84RR190 pKa = 11.84QGALEE195 pKa = 4.03RR196 pKa = 11.84MLVTT200 pKa = 4.93
MM1 pKa = 7.53LVTLQRR7 pKa = 11.84RR8 pKa = 11.84QGALQRR14 pKa = 11.84MLGSLQRR21 pKa = 11.84KK22 pKa = 9.15LGALQRR28 pKa = 11.84WLGALQRR35 pKa = 11.84RR36 pKa = 11.84QVALQRR42 pKa = 11.84MLGSLQRR49 pKa = 11.84MLRR52 pKa = 11.84ALQRR56 pKa = 11.84LLGALQRR63 pKa = 11.84RR64 pKa = 11.84QVALQRR70 pKa = 11.84RR71 pKa = 11.84QGASQRR77 pKa = 11.84WLGVLQRR84 pKa = 11.84WLGALQRR91 pKa = 11.84RR92 pKa = 11.84QVALQRR98 pKa = 11.84WLRR101 pKa = 11.84AVQRR105 pKa = 11.84MLGALQRR112 pKa = 11.84WLGALQRR119 pKa = 11.84WLEE122 pKa = 3.99ALQRR126 pKa = 11.84RR127 pKa = 11.84QGALQRR133 pKa = 11.84RR134 pKa = 11.84QGALQRR140 pKa = 11.84WPVTLQRR147 pKa = 11.84KK148 pKa = 8.14QVDD151 pKa = 4.12LQRR154 pKa = 11.84MLGSLQRR161 pKa = 11.84KK162 pKa = 9.15LGALQRR168 pKa = 11.84WLGALQRR175 pKa = 11.84MLSALQRR182 pKa = 11.84WLVTLQRR189 pKa = 11.84RR190 pKa = 11.84QGALEE195 pKa = 4.03RR196 pKa = 11.84MLVTT200 pKa = 4.93
Molecular weight: 23.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
37456558 |
8 |
5625 |
383.8 |
43.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.486 ± 0.008 | 1.951 ± 0.004 |
5.283 ± 0.005 | 6.489 ± 0.008 |
4.304 ± 0.005 | 6.174 ± 0.007 |
2.615 ± 0.003 | 5.451 ± 0.006 |
6.154 ± 0.008 | 9.549 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.49 ± 0.003 | 4.681 ± 0.006 |
4.853 ± 0.008 | 3.718 ± 0.005 |
5.433 ± 0.007 | 8.709 ± 0.009 |
4.939 ± 0.005 | 6.476 ± 0.007 |
1.352 ± 0.003 | 2.892 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
