
Wuhan insect virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
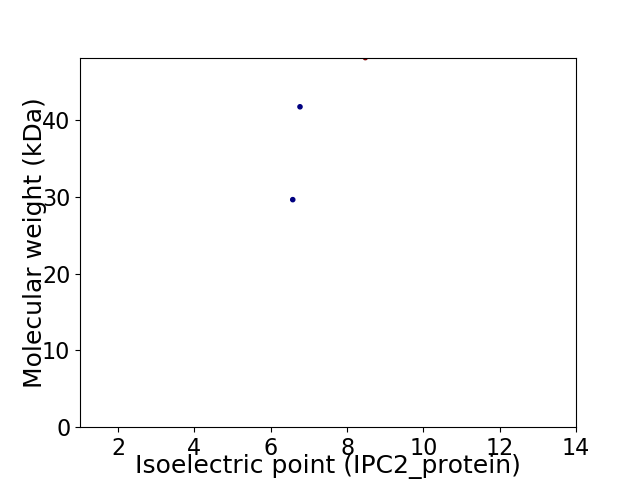
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGQ5|A0A1L3KGQ5_9VIRU Uncharacterized protein OS=Wuhan insect virus 20 OX=1923724 PE=4 SV=1
MM1 pKa = 7.76FDD3 pKa = 3.37QLLTASVRR11 pKa = 11.84VIRR14 pKa = 11.84DD15 pKa = 4.57FISFCYY21 pKa = 10.01NKK23 pKa = 10.3AGSIYY28 pKa = 10.38CSLKK32 pKa = 9.82KK33 pKa = 9.82WLWDD37 pKa = 3.57LQGKK41 pKa = 7.2FQQYY45 pKa = 10.63DD46 pKa = 3.49AFVDD50 pKa = 4.03LCYY53 pKa = 11.13GFMDD57 pKa = 4.14DD58 pKa = 4.05VEE60 pKa = 4.23EE61 pKa = 5.46AEE63 pKa = 4.24FEE65 pKa = 4.05RR66 pKa = 11.84LQVYY70 pKa = 9.72EE71 pKa = 4.4DD72 pKa = 3.67SLMEE76 pKa = 3.82EE77 pKa = 3.82SLARR81 pKa = 11.84KK82 pKa = 8.61EE83 pKa = 4.0LSRR86 pKa = 11.84ILALPVAGCWPMPTRR101 pKa = 11.84PPGSVNCEE109 pKa = 3.6EE110 pKa = 4.41YY111 pKa = 10.59GYY113 pKa = 11.08EE114 pKa = 4.01SALPADD120 pKa = 4.25HH121 pKa = 6.55QALIPEE127 pKa = 4.43EE128 pKa = 3.87NVQAVKK134 pKa = 10.09PVPLKK139 pKa = 10.38LQPKK143 pKa = 7.86TAVSQEE149 pKa = 3.77EE150 pKa = 4.6RR151 pKa = 11.84EE152 pKa = 4.2TAFLNFAAATAKK164 pKa = 10.07FDD166 pKa = 4.65QIKK169 pKa = 8.39EE170 pKa = 4.46TIKK173 pKa = 10.78DD174 pKa = 3.75LYY176 pKa = 11.06AEE178 pKa = 4.38EE179 pKa = 5.04RR180 pKa = 11.84GPTPFGRR187 pKa = 11.84WFGTLKK193 pKa = 10.34QRR195 pKa = 11.84MATVKK200 pKa = 8.99QAKK203 pKa = 9.07IRR205 pKa = 11.84RR206 pKa = 11.84EE207 pKa = 3.68RR208 pKa = 11.84SKK210 pKa = 11.01VYY212 pKa = 9.7AAKK215 pKa = 10.54VEE217 pKa = 3.9QEE219 pKa = 3.78MAYY222 pKa = 9.91KK223 pKa = 10.55EE224 pKa = 4.35NIPEE228 pKa = 4.17LLALTICVEE237 pKa = 3.96VDD239 pKa = 2.81TGEE242 pKa = 4.25PLPVKK247 pKa = 10.16KK248 pKa = 10.44DD249 pKa = 3.22EE250 pKa = 4.27QGAVRR255 pKa = 11.84AGFSPGTKK263 pKa = 9.16KK264 pKa = 10.89VLMRR268 pKa = 11.84QISSSPSDD276 pKa = 3.53RR277 pKa = 11.84KK278 pKa = 10.49DD279 pKa = 2.92AANWIRR285 pKa = 11.84HH286 pKa = 4.03YY287 pKa = 10.99VRR289 pKa = 11.84NKK291 pKa = 9.44NRR293 pKa = 11.84RR294 pKa = 11.84LSSDD298 pKa = 3.62EE299 pKa = 4.01VSHH302 pKa = 6.27ATIQRR307 pKa = 11.84YY308 pKa = 8.17VEE310 pKa = 4.34QICSDD315 pKa = 4.13LKK317 pKa = 11.23LDD319 pKa = 4.33LSSTTFLLDD328 pKa = 3.24KK329 pKa = 11.05ALMTVPVPTKK339 pKa = 10.35RR340 pKa = 11.84DD341 pKa = 3.43LKK343 pKa = 10.77SAMIVHH349 pKa = 6.39SPAACSLRR357 pKa = 11.84RR358 pKa = 11.84EE359 pKa = 4.1LAVLNSSVFF368 pKa = 3.34
MM1 pKa = 7.76FDD3 pKa = 3.37QLLTASVRR11 pKa = 11.84VIRR14 pKa = 11.84DD15 pKa = 4.57FISFCYY21 pKa = 10.01NKK23 pKa = 10.3AGSIYY28 pKa = 10.38CSLKK32 pKa = 9.82KK33 pKa = 9.82WLWDD37 pKa = 3.57LQGKK41 pKa = 7.2FQQYY45 pKa = 10.63DD46 pKa = 3.49AFVDD50 pKa = 4.03LCYY53 pKa = 11.13GFMDD57 pKa = 4.14DD58 pKa = 4.05VEE60 pKa = 4.23EE61 pKa = 5.46AEE63 pKa = 4.24FEE65 pKa = 4.05RR66 pKa = 11.84LQVYY70 pKa = 9.72EE71 pKa = 4.4DD72 pKa = 3.67SLMEE76 pKa = 3.82EE77 pKa = 3.82SLARR81 pKa = 11.84KK82 pKa = 8.61EE83 pKa = 4.0LSRR86 pKa = 11.84ILALPVAGCWPMPTRR101 pKa = 11.84PPGSVNCEE109 pKa = 3.6EE110 pKa = 4.41YY111 pKa = 10.59GYY113 pKa = 11.08EE114 pKa = 4.01SALPADD120 pKa = 4.25HH121 pKa = 6.55QALIPEE127 pKa = 4.43EE128 pKa = 3.87NVQAVKK134 pKa = 10.09PVPLKK139 pKa = 10.38LQPKK143 pKa = 7.86TAVSQEE149 pKa = 3.77EE150 pKa = 4.6RR151 pKa = 11.84EE152 pKa = 4.2TAFLNFAAATAKK164 pKa = 10.07FDD166 pKa = 4.65QIKK169 pKa = 8.39EE170 pKa = 4.46TIKK173 pKa = 10.78DD174 pKa = 3.75LYY176 pKa = 11.06AEE178 pKa = 4.38EE179 pKa = 5.04RR180 pKa = 11.84GPTPFGRR187 pKa = 11.84WFGTLKK193 pKa = 10.34QRR195 pKa = 11.84MATVKK200 pKa = 8.99QAKK203 pKa = 9.07IRR205 pKa = 11.84RR206 pKa = 11.84EE207 pKa = 3.68RR208 pKa = 11.84SKK210 pKa = 11.01VYY212 pKa = 9.7AAKK215 pKa = 10.54VEE217 pKa = 3.9QEE219 pKa = 3.78MAYY222 pKa = 9.91KK223 pKa = 10.55EE224 pKa = 4.35NIPEE228 pKa = 4.17LLALTICVEE237 pKa = 3.96VDD239 pKa = 2.81TGEE242 pKa = 4.25PLPVKK247 pKa = 10.16KK248 pKa = 10.44DD249 pKa = 3.22EE250 pKa = 4.27QGAVRR255 pKa = 11.84AGFSPGTKK263 pKa = 9.16KK264 pKa = 10.89VLMRR268 pKa = 11.84QISSSPSDD276 pKa = 3.53RR277 pKa = 11.84KK278 pKa = 10.49DD279 pKa = 2.92AANWIRR285 pKa = 11.84HH286 pKa = 4.03YY287 pKa = 10.99VRR289 pKa = 11.84NKK291 pKa = 9.44NRR293 pKa = 11.84RR294 pKa = 11.84LSSDD298 pKa = 3.62EE299 pKa = 4.01VSHH302 pKa = 6.27ATIQRR307 pKa = 11.84YY308 pKa = 8.17VEE310 pKa = 4.34QICSDD315 pKa = 4.13LKK317 pKa = 11.23LDD319 pKa = 4.33LSSTTFLLDD328 pKa = 3.24KK329 pKa = 11.05ALMTVPVPTKK339 pKa = 10.35RR340 pKa = 11.84DD341 pKa = 3.43LKK343 pKa = 10.77SAMIVHH349 pKa = 6.39SPAACSLRR357 pKa = 11.84RR358 pKa = 11.84EE359 pKa = 4.1LAVLNSSVFF368 pKa = 3.34
Molecular weight: 41.76 kDa
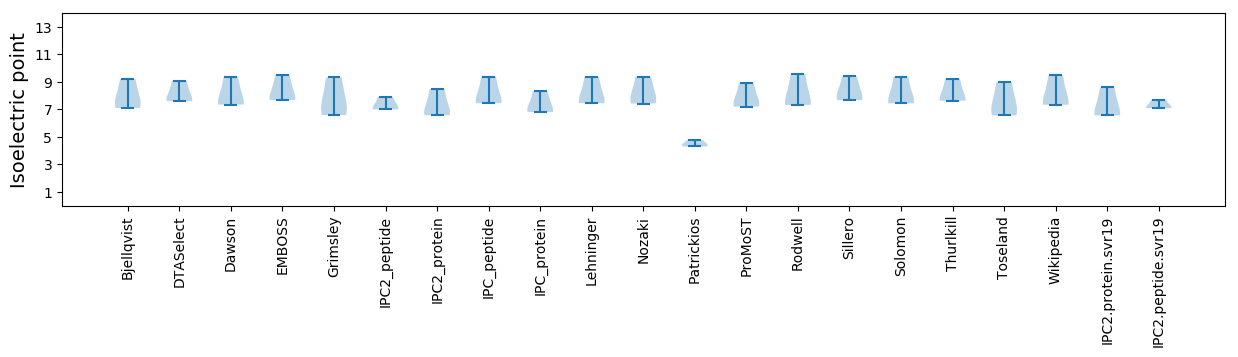
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGW1|A0A1L3KGW1_9VIRU RNA-directed RNA polymerase OS=Wuhan insect virus 20 OX=1923724 PE=4 SV=1
MM1 pKa = 7.53EE2 pKa = 5.62LANTYY7 pKa = 10.93SSGKK11 pKa = 7.82KK12 pKa = 8.91AQYY15 pKa = 10.17ISAVLNLRR23 pKa = 11.84KK24 pKa = 9.83RR25 pKa = 11.84PVEE28 pKa = 3.9QKK30 pKa = 10.8DD31 pKa = 3.75SYY33 pKa = 9.82VTAFLKK39 pKa = 9.99MEE41 pKa = 4.28KK42 pKa = 9.74HH43 pKa = 5.07WMCKK47 pKa = 9.89KK48 pKa = 9.69IAPRR52 pKa = 11.84LICPRR57 pKa = 11.84TKK59 pKa = 10.26RR60 pKa = 11.84YY61 pKa = 9.61NVEE64 pKa = 3.48LGRR67 pKa = 11.84RR68 pKa = 11.84LKK70 pKa = 10.88FNEE73 pKa = 4.33KK74 pKa = 10.43KK75 pKa = 10.46FMHH78 pKa = 7.14AIDD81 pKa = 4.38AVFGSPTVLSGYY93 pKa = 11.02DD94 pKa = 3.26NFNVGKK100 pKa = 10.1IIKK103 pKa = 9.88KK104 pKa = 9.94KK105 pKa = 7.63WDD107 pKa = 3.51KK108 pKa = 10.95FNEE111 pKa = 4.14PVAIGVDD118 pKa = 3.52ASRR121 pKa = 11.84FDD123 pKa = 3.45QHH125 pKa = 8.23VSEE128 pKa = 4.57QALRR132 pKa = 11.84WEE134 pKa = 3.84HH135 pKa = 6.57SIYY138 pKa = 11.16NKK140 pKa = 9.77VFHH143 pKa = 7.13DD144 pKa = 4.16PEE146 pKa = 5.19LALLLKK152 pKa = 9.58WQLTNYY158 pKa = 9.82VSLFVEE164 pKa = 4.47DD165 pKa = 4.33RR166 pKa = 11.84MLKK169 pKa = 10.42FKK171 pKa = 11.29VKK173 pKa = 8.36GHH175 pKa = 6.19RR176 pKa = 11.84MSGDD180 pKa = 3.0INTSMGNKK188 pKa = 10.21LIMCGMMHH196 pKa = 7.44KK197 pKa = 10.11YY198 pKa = 10.28IKK200 pKa = 10.5DD201 pKa = 3.54LGVKK205 pKa = 10.33AEE207 pKa = 4.27LCNNGDD213 pKa = 3.8DD214 pKa = 4.66CVIICEE220 pKa = 4.29RR221 pKa = 11.84SDD223 pKa = 3.18EE224 pKa = 4.23RR225 pKa = 11.84KK226 pKa = 9.78FDD228 pKa = 4.41GMQDD232 pKa = 2.65WFLNFGFNMVTEE244 pKa = 4.3EE245 pKa = 3.91PVYY248 pKa = 10.38EE249 pKa = 3.99IEE251 pKa = 5.23KK252 pKa = 11.17LEE254 pKa = 4.19FCQSKK259 pKa = 9.55PVSVGGKK266 pKa = 7.91YY267 pKa = 10.32RR268 pKa = 11.84MVRR271 pKa = 11.84RR272 pKa = 11.84PDD274 pKa = 4.06SISKK278 pKa = 10.28DD279 pKa = 3.55SHH281 pKa = 6.97SLLSMKK287 pKa = 10.31SRR289 pKa = 11.84EE290 pKa = 4.11DD291 pKa = 3.3TKK293 pKa = 11.55SFMSATAQCGLVLNSGVPILEE314 pKa = 4.12AFHH317 pKa = 7.12RR318 pKa = 11.84SMYY321 pKa = 10.23RR322 pKa = 11.84GSGYY326 pKa = 10.91KK327 pKa = 10.04KK328 pKa = 10.45ISDD331 pKa = 3.84AYY333 pKa = 8.2LKK335 pKa = 10.68KK336 pKa = 10.49VISYY340 pKa = 7.91GTDD343 pKa = 2.74EE344 pKa = 4.18RR345 pKa = 11.84LGTRR349 pKa = 11.84RR350 pKa = 11.84SWNDD354 pKa = 2.98EE355 pKa = 3.72PVTMEE360 pKa = 4.41NRR362 pKa = 11.84LSYY365 pKa = 9.82WKK367 pKa = 10.86AFGIDD372 pKa = 3.78PNTQEE377 pKa = 3.94LVEE380 pKa = 4.77RR381 pKa = 11.84YY382 pKa = 9.26FDD384 pKa = 3.93NLDD387 pKa = 3.17VCIEE391 pKa = 4.04SRR393 pKa = 11.84GVKK396 pKa = 10.11SLTPFLQSIVLNIPQHH412 pKa = 6.02PRR414 pKa = 11.84FCC416 pKa = 5.71
MM1 pKa = 7.53EE2 pKa = 5.62LANTYY7 pKa = 10.93SSGKK11 pKa = 7.82KK12 pKa = 8.91AQYY15 pKa = 10.17ISAVLNLRR23 pKa = 11.84KK24 pKa = 9.83RR25 pKa = 11.84PVEE28 pKa = 3.9QKK30 pKa = 10.8DD31 pKa = 3.75SYY33 pKa = 9.82VTAFLKK39 pKa = 9.99MEE41 pKa = 4.28KK42 pKa = 9.74HH43 pKa = 5.07WMCKK47 pKa = 9.89KK48 pKa = 9.69IAPRR52 pKa = 11.84LICPRR57 pKa = 11.84TKK59 pKa = 10.26RR60 pKa = 11.84YY61 pKa = 9.61NVEE64 pKa = 3.48LGRR67 pKa = 11.84RR68 pKa = 11.84LKK70 pKa = 10.88FNEE73 pKa = 4.33KK74 pKa = 10.43KK75 pKa = 10.46FMHH78 pKa = 7.14AIDD81 pKa = 4.38AVFGSPTVLSGYY93 pKa = 11.02DD94 pKa = 3.26NFNVGKK100 pKa = 10.1IIKK103 pKa = 9.88KK104 pKa = 9.94KK105 pKa = 7.63WDD107 pKa = 3.51KK108 pKa = 10.95FNEE111 pKa = 4.14PVAIGVDD118 pKa = 3.52ASRR121 pKa = 11.84FDD123 pKa = 3.45QHH125 pKa = 8.23VSEE128 pKa = 4.57QALRR132 pKa = 11.84WEE134 pKa = 3.84HH135 pKa = 6.57SIYY138 pKa = 11.16NKK140 pKa = 9.77VFHH143 pKa = 7.13DD144 pKa = 4.16PEE146 pKa = 5.19LALLLKK152 pKa = 9.58WQLTNYY158 pKa = 9.82VSLFVEE164 pKa = 4.47DD165 pKa = 4.33RR166 pKa = 11.84MLKK169 pKa = 10.42FKK171 pKa = 11.29VKK173 pKa = 8.36GHH175 pKa = 6.19RR176 pKa = 11.84MSGDD180 pKa = 3.0INTSMGNKK188 pKa = 10.21LIMCGMMHH196 pKa = 7.44KK197 pKa = 10.11YY198 pKa = 10.28IKK200 pKa = 10.5DD201 pKa = 3.54LGVKK205 pKa = 10.33AEE207 pKa = 4.27LCNNGDD213 pKa = 3.8DD214 pKa = 4.66CVIICEE220 pKa = 4.29RR221 pKa = 11.84SDD223 pKa = 3.18EE224 pKa = 4.23RR225 pKa = 11.84KK226 pKa = 9.78FDD228 pKa = 4.41GMQDD232 pKa = 2.65WFLNFGFNMVTEE244 pKa = 4.3EE245 pKa = 3.91PVYY248 pKa = 10.38EE249 pKa = 3.99IEE251 pKa = 5.23KK252 pKa = 11.17LEE254 pKa = 4.19FCQSKK259 pKa = 9.55PVSVGGKK266 pKa = 7.91YY267 pKa = 10.32RR268 pKa = 11.84MVRR271 pKa = 11.84RR272 pKa = 11.84PDD274 pKa = 4.06SISKK278 pKa = 10.28DD279 pKa = 3.55SHH281 pKa = 6.97SLLSMKK287 pKa = 10.31SRR289 pKa = 11.84EE290 pKa = 4.11DD291 pKa = 3.3TKK293 pKa = 11.55SFMSATAQCGLVLNSGVPILEE314 pKa = 4.12AFHH317 pKa = 7.12RR318 pKa = 11.84SMYY321 pKa = 10.23RR322 pKa = 11.84GSGYY326 pKa = 10.91KK327 pKa = 10.04KK328 pKa = 10.45ISDD331 pKa = 3.84AYY333 pKa = 8.2LKK335 pKa = 10.68KK336 pKa = 10.49VISYY340 pKa = 7.91GTDD343 pKa = 2.74EE344 pKa = 4.18RR345 pKa = 11.84LGTRR349 pKa = 11.84RR350 pKa = 11.84SWNDD354 pKa = 2.98EE355 pKa = 3.72PVTMEE360 pKa = 4.41NRR362 pKa = 11.84LSYY365 pKa = 9.82WKK367 pKa = 10.86AFGIDD372 pKa = 3.78PNTQEE377 pKa = 3.94LVEE380 pKa = 4.77RR381 pKa = 11.84YY382 pKa = 9.26FDD384 pKa = 3.93NLDD387 pKa = 3.17VCIEE391 pKa = 4.04SRR393 pKa = 11.84GVKK396 pKa = 10.11SLTPFLQSIVLNIPQHH412 pKa = 6.02PRR414 pKa = 11.84FCC416 pKa = 5.71
Molecular weight: 48.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1050 |
266 |
416 |
350.0 |
39.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.476 ± 1.355 | 1.905 ± 0.38 |
5.81 ± 0.209 | 6.667 ± 0.68 |
4.381 ± 0.325 | 5.238 ± 0.773 |
2.095 ± 0.458 | 4.667 ± 0.322 |
7.81 ± 0.791 | 7.714 ± 1.103 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.952 ± 0.554 | 4.19 ± 0.77 |
4.476 ± 0.481 | 3.524 ± 0.522 |
6.19 ± 0.194 | 8.762 ± 1.077 |
4.381 ± 0.525 | 7.429 ± 0.29 |
1.524 ± 0.085 | 3.81 ± 0.276 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
