
Homalodisca vitripennis reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Phytoreovirus; unclassified Phytoreovirus
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
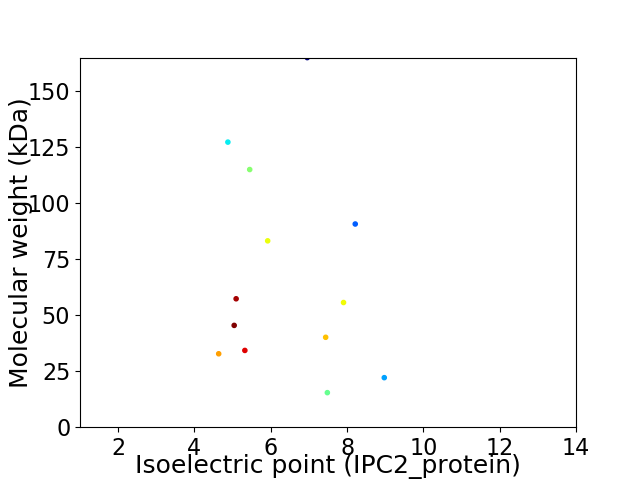
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C1JAW7|C1JAW7_9REOV Suppressor of RNA-mediated gene silencing OS=Homalodisca vitripennis reovirus OX=411854 PE=3 SV=1
MM1 pKa = 7.63SGQKK5 pKa = 10.2LQDD8 pKa = 3.0GTAIKK13 pKa = 10.13RR14 pKa = 11.84INDD17 pKa = 4.76AIQIFQKK24 pKa = 10.34YY25 pKa = 5.97QTGEE29 pKa = 3.92VSKK32 pKa = 11.13SSSNHH37 pKa = 5.32LNQLRR42 pKa = 11.84NIRR45 pKa = 11.84MSVGLAWPVILKK57 pKa = 10.18YY58 pKa = 10.8CFLHH62 pKa = 6.35VSSHH66 pKa = 6.02CGVVRR71 pKa = 11.84FLTDD75 pKa = 3.59LACTVKK81 pKa = 10.89VGVFPIISSIGDD93 pKa = 3.89FDD95 pKa = 4.79PFDD98 pKa = 4.29DD99 pKa = 4.34VGLIFSKK106 pKa = 10.72SCIALKK112 pKa = 10.51ISDD115 pKa = 4.38SSFLTPPDD123 pKa = 3.4SFASKK128 pKa = 9.12LTLFFSAMSVNARR141 pKa = 11.84VEE143 pKa = 3.71TDD145 pKa = 3.12MLHH148 pKa = 6.41EE149 pKa = 5.05IEE151 pKa = 5.74DD152 pKa = 4.15RR153 pKa = 11.84YY154 pKa = 9.08TYY156 pKa = 8.94RR157 pKa = 11.84TGVIGEE163 pKa = 4.22LLSVYY168 pKa = 10.25MNATLNLDD176 pKa = 3.54DD177 pKa = 4.17WVVNLDD183 pKa = 4.01EE184 pKa = 5.49LSQPTRR190 pKa = 11.84AQMLTLLAQNRR201 pKa = 11.84NMISNEE207 pKa = 2.74IRR209 pKa = 11.84NYY211 pKa = 9.44VNKK214 pKa = 10.4NLDD217 pKa = 3.46HH218 pKa = 6.85PSEE221 pKa = 4.23HH222 pKa = 6.56LAAVAKK228 pKa = 10.2KK229 pKa = 10.27YY230 pKa = 8.8NQEE233 pKa = 3.54WDD235 pKa = 3.03IGQPTPNLLPVTSATEE251 pKa = 3.66IDD253 pKa = 4.2GPEE256 pKa = 4.15SDD258 pKa = 4.71TEE260 pKa = 4.16SDD262 pKa = 3.88SYY264 pKa = 11.5SASTDD269 pKa = 3.01LDD271 pKa = 3.49ASLRR275 pKa = 11.84AVSSFPTRR283 pKa = 11.84DD284 pKa = 3.47DD285 pKa = 4.26EE286 pKa = 5.78IEE288 pKa = 4.11FSLDD292 pKa = 3.4DD293 pKa = 3.9LL294 pKa = 4.86
MM1 pKa = 7.63SGQKK5 pKa = 10.2LQDD8 pKa = 3.0GTAIKK13 pKa = 10.13RR14 pKa = 11.84INDD17 pKa = 4.76AIQIFQKK24 pKa = 10.34YY25 pKa = 5.97QTGEE29 pKa = 3.92VSKK32 pKa = 11.13SSSNHH37 pKa = 5.32LNQLRR42 pKa = 11.84NIRR45 pKa = 11.84MSVGLAWPVILKK57 pKa = 10.18YY58 pKa = 10.8CFLHH62 pKa = 6.35VSSHH66 pKa = 6.02CGVVRR71 pKa = 11.84FLTDD75 pKa = 3.59LACTVKK81 pKa = 10.89VGVFPIISSIGDD93 pKa = 3.89FDD95 pKa = 4.79PFDD98 pKa = 4.29DD99 pKa = 4.34VGLIFSKK106 pKa = 10.72SCIALKK112 pKa = 10.51ISDD115 pKa = 4.38SSFLTPPDD123 pKa = 3.4SFASKK128 pKa = 9.12LTLFFSAMSVNARR141 pKa = 11.84VEE143 pKa = 3.71TDD145 pKa = 3.12MLHH148 pKa = 6.41EE149 pKa = 5.05IEE151 pKa = 5.74DD152 pKa = 4.15RR153 pKa = 11.84YY154 pKa = 9.08TYY156 pKa = 8.94RR157 pKa = 11.84TGVIGEE163 pKa = 4.22LLSVYY168 pKa = 10.25MNATLNLDD176 pKa = 3.54DD177 pKa = 4.17WVVNLDD183 pKa = 4.01EE184 pKa = 5.49LSQPTRR190 pKa = 11.84AQMLTLLAQNRR201 pKa = 11.84NMISNEE207 pKa = 2.74IRR209 pKa = 11.84NYY211 pKa = 9.44VNKK214 pKa = 10.4NLDD217 pKa = 3.46HH218 pKa = 6.85PSEE221 pKa = 4.23HH222 pKa = 6.56LAAVAKK228 pKa = 10.2KK229 pKa = 10.27YY230 pKa = 8.8NQEE233 pKa = 3.54WDD235 pKa = 3.03IGQPTPNLLPVTSATEE251 pKa = 3.66IDD253 pKa = 4.2GPEE256 pKa = 4.15SDD258 pKa = 4.71TEE260 pKa = 4.16SDD262 pKa = 3.88SYY264 pKa = 11.5SASTDD269 pKa = 3.01LDD271 pKa = 3.49ASLRR275 pKa = 11.84AVSSFPTRR283 pKa = 11.84DD284 pKa = 3.47DD285 pKa = 4.26EE286 pKa = 5.78IEE288 pKa = 4.11FSLDD292 pKa = 3.4DD293 pKa = 3.9LL294 pKa = 4.86
Molecular weight: 32.74 kDa
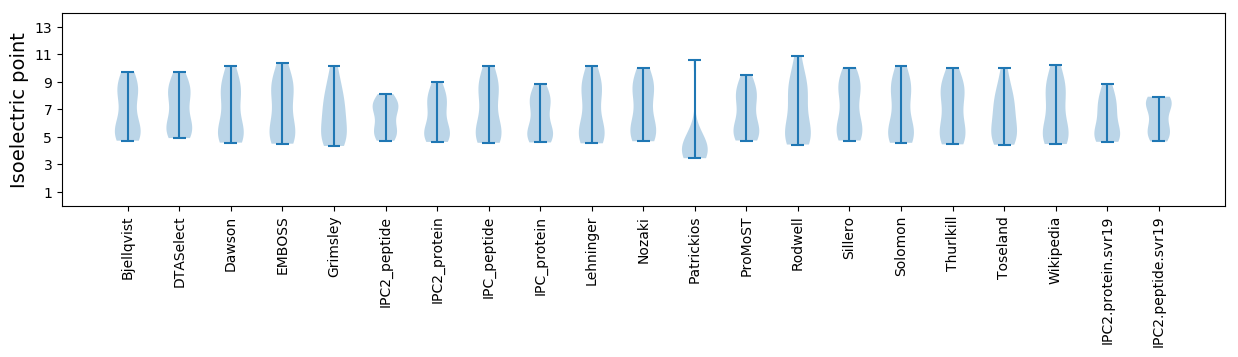
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C1JAW9|C1JAW9_9REOV Uncharacterized protein OS=Homalodisca vitripennis reovirus OX=411854 PE=4 SV=1
MM1 pKa = 7.26NSSVSHH7 pKa = 6.37VGTITIAPDD16 pKa = 3.24KK17 pKa = 10.82DD18 pKa = 3.34SEE20 pKa = 4.34QIYY23 pKa = 10.13KK24 pKa = 10.31RR25 pKa = 11.84KK26 pKa = 9.91KK27 pKa = 8.6LQLPATLKK35 pKa = 10.47INYY38 pKa = 9.29IKK40 pKa = 10.28DD41 pKa = 3.01AKK43 pKa = 10.71YY44 pKa = 10.22RR45 pKa = 11.84ICACPSDD52 pKa = 3.77SHH54 pKa = 8.89SIDD57 pKa = 3.78DD58 pKa = 4.55CPLGGLPQSFVAHH71 pKa = 5.6KK72 pKa = 8.13TLWLSSLLHH81 pKa = 6.4SSSTSANSRR90 pKa = 11.84KK91 pKa = 9.76AVLPRR96 pKa = 11.84IKK98 pKa = 10.02QSKK101 pKa = 9.47FRR103 pKa = 11.84LTDD106 pKa = 2.99TSRR109 pKa = 11.84KK110 pKa = 9.3VKK112 pKa = 10.55SKK114 pKa = 11.33SEE116 pKa = 4.03DD117 pKa = 3.13ATKK120 pKa = 10.91LEE122 pKa = 4.24RR123 pKa = 11.84ASEE126 pKa = 4.03KK127 pKa = 10.43QLEE130 pKa = 4.53KK131 pKa = 10.44KK132 pKa = 10.4SKK134 pKa = 9.52SSEE137 pKa = 3.99VQEE140 pKa = 4.19SSEE143 pKa = 4.01EE144 pKa = 4.4SEE146 pKa = 4.86GEE148 pKa = 4.18TSHH151 pKa = 6.89SRR153 pKa = 11.84LKK155 pKa = 10.48RR156 pKa = 11.84HH157 pKa = 5.92QKK159 pKa = 9.68SEE161 pKa = 3.95KK162 pKa = 9.76KK163 pKa = 10.05GRR165 pKa = 11.84KK166 pKa = 8.74HH167 pKa = 6.91DD168 pKa = 4.13SKK170 pKa = 10.92STLSTEE176 pKa = 4.24EE177 pKa = 3.79VTKK180 pKa = 10.77LVAAITTEE188 pKa = 4.0ALSKK192 pKa = 9.79LTRR195 pKa = 11.84LKK197 pKa = 10.92DD198 pKa = 3.29
MM1 pKa = 7.26NSSVSHH7 pKa = 6.37VGTITIAPDD16 pKa = 3.24KK17 pKa = 10.82DD18 pKa = 3.34SEE20 pKa = 4.34QIYY23 pKa = 10.13KK24 pKa = 10.31RR25 pKa = 11.84KK26 pKa = 9.91KK27 pKa = 8.6LQLPATLKK35 pKa = 10.47INYY38 pKa = 9.29IKK40 pKa = 10.28DD41 pKa = 3.01AKK43 pKa = 10.71YY44 pKa = 10.22RR45 pKa = 11.84ICACPSDD52 pKa = 3.77SHH54 pKa = 8.89SIDD57 pKa = 3.78DD58 pKa = 4.55CPLGGLPQSFVAHH71 pKa = 5.6KK72 pKa = 8.13TLWLSSLLHH81 pKa = 6.4SSSTSANSRR90 pKa = 11.84KK91 pKa = 9.76AVLPRR96 pKa = 11.84IKK98 pKa = 10.02QSKK101 pKa = 9.47FRR103 pKa = 11.84LTDD106 pKa = 2.99TSRR109 pKa = 11.84KK110 pKa = 9.3VKK112 pKa = 10.55SKK114 pKa = 11.33SEE116 pKa = 4.03DD117 pKa = 3.13ATKK120 pKa = 10.91LEE122 pKa = 4.24RR123 pKa = 11.84ASEE126 pKa = 4.03KK127 pKa = 10.43QLEE130 pKa = 4.53KK131 pKa = 10.44KK132 pKa = 10.4SKK134 pKa = 9.52SSEE137 pKa = 3.99VQEE140 pKa = 4.19SSEE143 pKa = 4.01EE144 pKa = 4.4SEE146 pKa = 4.86GEE148 pKa = 4.18TSHH151 pKa = 6.89SRR153 pKa = 11.84LKK155 pKa = 10.48RR156 pKa = 11.84HH157 pKa = 5.92QKK159 pKa = 9.68SEE161 pKa = 3.95KK162 pKa = 9.76KK163 pKa = 10.05GRR165 pKa = 11.84KK166 pKa = 8.74HH167 pKa = 6.91DD168 pKa = 4.13SKK170 pKa = 10.92STLSTEE176 pKa = 4.24EE177 pKa = 3.79VTKK180 pKa = 10.77LVAAITTEE188 pKa = 4.0ALSKK192 pKa = 9.79LTRR195 pKa = 11.84LKK197 pKa = 10.92DD198 pKa = 3.29
Molecular weight: 22.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7927 |
135 |
1460 |
609.8 |
67.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.812 ± 0.425 | 1.173 ± 0.182 |
6.068 ± 0.221 | 4.642 ± 0.429 |
4.062 ± 0.176 | 5.097 ± 0.296 |
1.791 ± 0.203 | 7.279 ± 0.305 |
5.853 ± 0.632 | 9.07 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.813 ± 0.183 | 5.639 ± 0.268 |
4.113 ± 0.243 | 3.141 ± 0.178 |
4.188 ± 0.258 | 9.461 ± 0.525 |
7.039 ± 0.42 | 6.825 ± 0.315 |
0.82 ± 0.127 | 4.113 ± 0.354 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
