
Hubei permutotetra-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
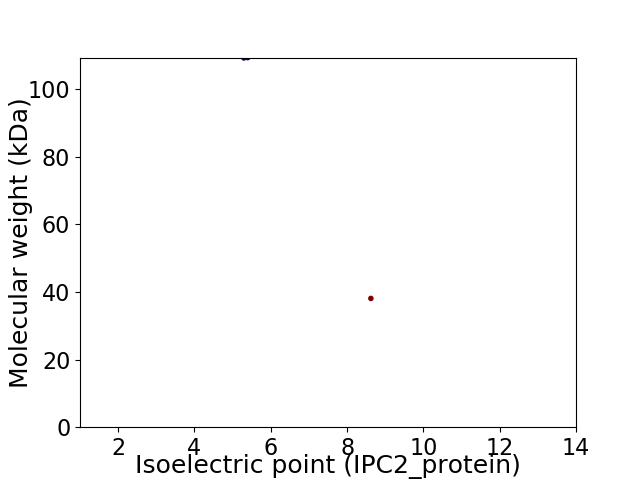
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHZ9|A0A1L3KHZ9_9VIRU RdRp OS=Hubei permutotetra-like virus 10 OX=1923074 PE=4 SV=1
MM1 pKa = 7.51EE2 pKa = 6.1NIMLQRR8 pKa = 11.84QDD10 pKa = 3.47YY11 pKa = 10.86NRR13 pKa = 11.84LQKK16 pKa = 10.46IIATAPRR23 pKa = 11.84TPAPFKK29 pKa = 11.2NPVEE33 pKa = 4.08LEE35 pKa = 4.1IEE37 pKa = 4.08EE38 pKa = 4.48STLKK42 pKa = 10.79RR43 pKa = 11.84LTRR46 pKa = 11.84DD47 pKa = 2.85IPVQGTLDD55 pKa = 3.52LSFLVKK61 pKa = 10.51GSEE64 pKa = 4.24VPLSPEE70 pKa = 3.7HH71 pKa = 6.51TPGLNQKK78 pKa = 9.75GAFVSVNPVFKK89 pKa = 10.05PASRR93 pKa = 11.84DD94 pKa = 3.37RR95 pKa = 11.84KK96 pKa = 9.88VVGAVLRR103 pKa = 11.84RR104 pKa = 11.84GGRR107 pKa = 11.84VEE109 pKa = 5.65DD110 pKa = 3.82IEE112 pKa = 4.25KK113 pKa = 10.68LIYY116 pKa = 7.88TTGSKK121 pKa = 10.3SGFSEE126 pKa = 4.82RR127 pKa = 11.84LKK129 pKa = 10.88LWATARR135 pKa = 11.84AQNPEE140 pKa = 4.08TEE142 pKa = 4.21LKK144 pKa = 10.57EE145 pKa = 4.04RR146 pKa = 11.84LLNAKK151 pKa = 10.12DD152 pKa = 3.84IIRR155 pKa = 11.84KK156 pKa = 8.97RR157 pKa = 11.84LPVRR161 pKa = 11.84EE162 pKa = 4.15KK163 pKa = 11.03LLPDD167 pKa = 3.62WNATVEE173 pKa = 4.16EE174 pKa = 4.76FVAGVTVNKK183 pKa = 10.0SSSAGPPLYY192 pKa = 10.01RR193 pKa = 11.84PKK195 pKa = 10.52YY196 pKa = 8.31QCTDD200 pKa = 2.83EE201 pKa = 4.32YY202 pKa = 10.64MDD204 pKa = 5.05IISEE208 pKa = 4.04IMKK211 pKa = 10.28EE212 pKa = 4.23GNQDD216 pKa = 3.06HH217 pKa = 5.88LTEE220 pKa = 4.08YY221 pKa = 11.18LEE223 pKa = 4.21GRR225 pKa = 11.84EE226 pKa = 4.01EE227 pKa = 5.41FMIAQCKK234 pKa = 9.1NKK236 pKa = 9.4QDD238 pKa = 3.78RR239 pKa = 11.84YY240 pKa = 10.4PPDD243 pKa = 3.63KK244 pKa = 10.67LDD246 pKa = 3.54EE247 pKa = 4.1KK248 pKa = 10.08TRR250 pKa = 11.84PYY252 pKa = 11.16FNFCFPLQFYY262 pKa = 9.68FSALVQPKK270 pKa = 6.9THH272 pKa = 6.65CSVLFTEE279 pKa = 6.47SEE281 pKa = 4.45DD282 pKa = 3.12SWNAYY287 pKa = 8.18GFSWANGGGEE297 pKa = 5.47KK298 pKa = 10.13FFKK301 pKa = 9.73WMKK304 pKa = 9.0SCRR307 pKa = 11.84EE308 pKa = 4.1KK309 pKa = 10.66EE310 pKa = 4.25VKK312 pKa = 10.26LAVYY316 pKa = 10.32GDD318 pKa = 4.05DD319 pKa = 3.39VFLVKK324 pKa = 10.51RR325 pKa = 11.84EE326 pKa = 3.91GGLLKK331 pKa = 10.68ACAPDD336 pKa = 4.06FRR338 pKa = 11.84SMDD341 pKa = 3.31GSVHH345 pKa = 6.67KK346 pKa = 10.64KK347 pKa = 9.38DD348 pKa = 5.06AEE350 pKa = 4.3WFCEE354 pKa = 3.74DD355 pKa = 4.65LIEE358 pKa = 4.85AFEE361 pKa = 4.05RR362 pKa = 11.84QYY364 pKa = 11.73GKK366 pKa = 10.7NNFWSYY372 pKa = 10.69FGKK375 pKa = 10.44LLSRR379 pKa = 11.84SLVGAKK385 pKa = 9.99FLVDD389 pKa = 4.12GPQAYY394 pKa = 10.48VNEE397 pKa = 4.4TGLMTGCVGTTGVDD411 pKa = 3.42TQKK414 pKa = 11.25SVVAYY419 pKa = 10.37ACLLDD424 pKa = 3.69KK425 pKa = 11.37AEE427 pKa = 4.13QKK429 pKa = 10.87EE430 pKa = 3.83IDD432 pKa = 4.32LEE434 pKa = 4.55DD435 pKa = 4.56GEE437 pKa = 5.19AVSTYY442 pKa = 9.72FRR444 pKa = 11.84QKK446 pKa = 9.87FGLIVKK452 pKa = 9.42EE453 pKa = 4.18NTYY456 pKa = 7.71EE457 pKa = 3.94WKK459 pKa = 10.56VVEE462 pKa = 4.38EE463 pKa = 3.99EE464 pKa = 4.33LEE466 pKa = 4.18EE467 pKa = 5.19GEE469 pKa = 4.6VPVDD473 pKa = 4.43LEE475 pKa = 4.06FLGVKK480 pKa = 10.22LKK482 pKa = 9.99MRR484 pKa = 11.84LGKK487 pKa = 9.99EE488 pKa = 3.63RR489 pKa = 11.84LEE491 pKa = 3.9CVPFKK496 pKa = 11.0EE497 pKa = 4.4EE498 pKa = 3.84NDD500 pKa = 3.81LVSLISNARR509 pKa = 11.84IPPEE513 pKa = 3.91AKK515 pKa = 10.02GRR517 pKa = 11.84ATIKK521 pKa = 10.51DD522 pKa = 3.28RR523 pKa = 11.84YY524 pKa = 8.96LFDD527 pKa = 3.25MARR530 pKa = 11.84GYY532 pKa = 10.12MITGAFHH539 pKa = 7.19SEE541 pKa = 4.4VVWNICCDD549 pKa = 4.3LIEE552 pKa = 4.39GTKK555 pKa = 9.88TEE557 pKa = 4.24IVCQRR562 pKa = 11.84VQSGKK567 pKa = 10.07KK568 pKa = 9.72VEE570 pKa = 4.12VAPGKK575 pKa = 9.45WEE577 pKa = 3.9HH578 pKa = 6.36TGQSPEE584 pKa = 4.14LEE586 pKa = 4.06QLVGSDD592 pKa = 4.07FQWPTSDD599 pKa = 3.05GWPSQEE605 pKa = 3.84FCLDD609 pKa = 3.53VYY611 pKa = 10.93LSPDD615 pKa = 3.25NKK617 pKa = 10.32IGGEE621 pKa = 4.11WYY623 pKa = 10.13SCVPSLEE630 pKa = 4.1EE631 pKa = 4.17DD632 pKa = 3.58LKK634 pKa = 11.07KK635 pKa = 10.71LRR637 pKa = 11.84SKK639 pKa = 10.61RR640 pKa = 11.84VTMEE644 pKa = 3.55AAKK647 pKa = 10.57LKK649 pKa = 9.75EE650 pKa = 4.05QGPKK654 pKa = 9.92FDD656 pKa = 3.94WALDD660 pKa = 3.46TDD662 pKa = 4.41LEE664 pKa = 4.52EE665 pKa = 5.04KK666 pKa = 10.59ASQLEE671 pKa = 3.91AVRR674 pKa = 11.84DD675 pKa = 3.98SKK677 pKa = 10.97IDD679 pKa = 3.4PRR681 pKa = 11.84VDD683 pKa = 3.1RR684 pKa = 11.84EE685 pKa = 4.3GYY687 pKa = 10.49LRR689 pKa = 11.84DD690 pKa = 3.62LVKK693 pKa = 10.42EE694 pKa = 4.15RR695 pKa = 11.84PLMRR699 pKa = 11.84PAKK702 pKa = 9.59QPKK705 pKa = 9.62NFLRR709 pKa = 11.84LKK711 pKa = 9.05GQEE714 pKa = 4.15SRR716 pKa = 11.84SKK718 pKa = 10.4DD719 pKa = 3.4QQIGDD724 pKa = 3.98FLEE727 pKa = 4.53KK728 pKa = 10.33FSSVLEE734 pKa = 4.03EE735 pKa = 5.44LPDD738 pKa = 3.48QMLEE742 pKa = 4.22LVVDD746 pKa = 4.55SPPGYY751 pKa = 9.85YY752 pKa = 10.02SRR754 pKa = 11.84VMQGKK759 pKa = 8.61GWRR762 pKa = 11.84PNPDD766 pKa = 2.6RR767 pKa = 11.84KK768 pKa = 9.69VWEE771 pKa = 4.5RR772 pKa = 11.84GSPVPVNPHH781 pKa = 3.23VWGRR785 pKa = 11.84DD786 pKa = 3.01ILMTLPDD793 pKa = 5.62FIPTEE798 pKa = 3.88KK799 pKa = 9.69KK800 pKa = 8.5TVRR803 pKa = 11.84KK804 pKa = 9.45EE805 pKa = 4.04CEE807 pKa = 3.61EE808 pKa = 4.4DD809 pKa = 3.81KK810 pKa = 10.05PTEE813 pKa = 4.19PFTLVPEE820 pKa = 4.3YY821 pKa = 11.19VKK823 pKa = 10.88LLPNVKK829 pKa = 8.89MDD831 pKa = 5.33AISLVTGSFSKK842 pKa = 10.99AGDD845 pKa = 3.16ILTTSNEE852 pKa = 4.11VLRR855 pKa = 11.84LSPDD859 pKa = 2.66SWIKK863 pKa = 9.66TEE865 pKa = 3.7VRR867 pKa = 11.84VRR869 pKa = 11.84NGTGIVARR877 pKa = 11.84AYY879 pKa = 10.31AKK881 pKa = 10.21SGKK884 pKa = 9.11AAKK887 pKa = 9.66EE888 pKa = 3.72MCFEE892 pKa = 4.56NMLEE896 pKa = 3.91QLKK899 pKa = 10.16EE900 pKa = 4.14RR901 pKa = 11.84EE902 pKa = 4.03RR903 pKa = 11.84QPTSFKK909 pKa = 10.78QQTEE913 pKa = 3.86EE914 pKa = 3.97DD915 pKa = 4.12GNRR918 pKa = 11.84STEE921 pKa = 4.0SEE923 pKa = 4.13AAAEE927 pKa = 4.28TQTTEE932 pKa = 4.03RR933 pKa = 11.84EE934 pKa = 4.19EE935 pKa = 4.28EE936 pKa = 3.95NGEE939 pKa = 4.16EE940 pKa = 4.19LVQDD944 pKa = 3.5VCEE947 pKa = 4.17VRR949 pKa = 11.84EE950 pKa = 4.32GTGGGDD956 pKa = 3.45GGVHH960 pKa = 6.6LL961 pKa = 5.16
MM1 pKa = 7.51EE2 pKa = 6.1NIMLQRR8 pKa = 11.84QDD10 pKa = 3.47YY11 pKa = 10.86NRR13 pKa = 11.84LQKK16 pKa = 10.46IIATAPRR23 pKa = 11.84TPAPFKK29 pKa = 11.2NPVEE33 pKa = 4.08LEE35 pKa = 4.1IEE37 pKa = 4.08EE38 pKa = 4.48STLKK42 pKa = 10.79RR43 pKa = 11.84LTRR46 pKa = 11.84DD47 pKa = 2.85IPVQGTLDD55 pKa = 3.52LSFLVKK61 pKa = 10.51GSEE64 pKa = 4.24VPLSPEE70 pKa = 3.7HH71 pKa = 6.51TPGLNQKK78 pKa = 9.75GAFVSVNPVFKK89 pKa = 10.05PASRR93 pKa = 11.84DD94 pKa = 3.37RR95 pKa = 11.84KK96 pKa = 9.88VVGAVLRR103 pKa = 11.84RR104 pKa = 11.84GGRR107 pKa = 11.84VEE109 pKa = 5.65DD110 pKa = 3.82IEE112 pKa = 4.25KK113 pKa = 10.68LIYY116 pKa = 7.88TTGSKK121 pKa = 10.3SGFSEE126 pKa = 4.82RR127 pKa = 11.84LKK129 pKa = 10.88LWATARR135 pKa = 11.84AQNPEE140 pKa = 4.08TEE142 pKa = 4.21LKK144 pKa = 10.57EE145 pKa = 4.04RR146 pKa = 11.84LLNAKK151 pKa = 10.12DD152 pKa = 3.84IIRR155 pKa = 11.84KK156 pKa = 8.97RR157 pKa = 11.84LPVRR161 pKa = 11.84EE162 pKa = 4.15KK163 pKa = 11.03LLPDD167 pKa = 3.62WNATVEE173 pKa = 4.16EE174 pKa = 4.76FVAGVTVNKK183 pKa = 10.0SSSAGPPLYY192 pKa = 10.01RR193 pKa = 11.84PKK195 pKa = 10.52YY196 pKa = 8.31QCTDD200 pKa = 2.83EE201 pKa = 4.32YY202 pKa = 10.64MDD204 pKa = 5.05IISEE208 pKa = 4.04IMKK211 pKa = 10.28EE212 pKa = 4.23GNQDD216 pKa = 3.06HH217 pKa = 5.88LTEE220 pKa = 4.08YY221 pKa = 11.18LEE223 pKa = 4.21GRR225 pKa = 11.84EE226 pKa = 4.01EE227 pKa = 5.41FMIAQCKK234 pKa = 9.1NKK236 pKa = 9.4QDD238 pKa = 3.78RR239 pKa = 11.84YY240 pKa = 10.4PPDD243 pKa = 3.63KK244 pKa = 10.67LDD246 pKa = 3.54EE247 pKa = 4.1KK248 pKa = 10.08TRR250 pKa = 11.84PYY252 pKa = 11.16FNFCFPLQFYY262 pKa = 9.68FSALVQPKK270 pKa = 6.9THH272 pKa = 6.65CSVLFTEE279 pKa = 6.47SEE281 pKa = 4.45DD282 pKa = 3.12SWNAYY287 pKa = 8.18GFSWANGGGEE297 pKa = 5.47KK298 pKa = 10.13FFKK301 pKa = 9.73WMKK304 pKa = 9.0SCRR307 pKa = 11.84EE308 pKa = 4.1KK309 pKa = 10.66EE310 pKa = 4.25VKK312 pKa = 10.26LAVYY316 pKa = 10.32GDD318 pKa = 4.05DD319 pKa = 3.39VFLVKK324 pKa = 10.51RR325 pKa = 11.84EE326 pKa = 3.91GGLLKK331 pKa = 10.68ACAPDD336 pKa = 4.06FRR338 pKa = 11.84SMDD341 pKa = 3.31GSVHH345 pKa = 6.67KK346 pKa = 10.64KK347 pKa = 9.38DD348 pKa = 5.06AEE350 pKa = 4.3WFCEE354 pKa = 3.74DD355 pKa = 4.65LIEE358 pKa = 4.85AFEE361 pKa = 4.05RR362 pKa = 11.84QYY364 pKa = 11.73GKK366 pKa = 10.7NNFWSYY372 pKa = 10.69FGKK375 pKa = 10.44LLSRR379 pKa = 11.84SLVGAKK385 pKa = 9.99FLVDD389 pKa = 4.12GPQAYY394 pKa = 10.48VNEE397 pKa = 4.4TGLMTGCVGTTGVDD411 pKa = 3.42TQKK414 pKa = 11.25SVVAYY419 pKa = 10.37ACLLDD424 pKa = 3.69KK425 pKa = 11.37AEE427 pKa = 4.13QKK429 pKa = 10.87EE430 pKa = 3.83IDD432 pKa = 4.32LEE434 pKa = 4.55DD435 pKa = 4.56GEE437 pKa = 5.19AVSTYY442 pKa = 9.72FRR444 pKa = 11.84QKK446 pKa = 9.87FGLIVKK452 pKa = 9.42EE453 pKa = 4.18NTYY456 pKa = 7.71EE457 pKa = 3.94WKK459 pKa = 10.56VVEE462 pKa = 4.38EE463 pKa = 3.99EE464 pKa = 4.33LEE466 pKa = 4.18EE467 pKa = 5.19GEE469 pKa = 4.6VPVDD473 pKa = 4.43LEE475 pKa = 4.06FLGVKK480 pKa = 10.22LKK482 pKa = 9.99MRR484 pKa = 11.84LGKK487 pKa = 9.99EE488 pKa = 3.63RR489 pKa = 11.84LEE491 pKa = 3.9CVPFKK496 pKa = 11.0EE497 pKa = 4.4EE498 pKa = 3.84NDD500 pKa = 3.81LVSLISNARR509 pKa = 11.84IPPEE513 pKa = 3.91AKK515 pKa = 10.02GRR517 pKa = 11.84ATIKK521 pKa = 10.51DD522 pKa = 3.28RR523 pKa = 11.84YY524 pKa = 8.96LFDD527 pKa = 3.25MARR530 pKa = 11.84GYY532 pKa = 10.12MITGAFHH539 pKa = 7.19SEE541 pKa = 4.4VVWNICCDD549 pKa = 4.3LIEE552 pKa = 4.39GTKK555 pKa = 9.88TEE557 pKa = 4.24IVCQRR562 pKa = 11.84VQSGKK567 pKa = 10.07KK568 pKa = 9.72VEE570 pKa = 4.12VAPGKK575 pKa = 9.45WEE577 pKa = 3.9HH578 pKa = 6.36TGQSPEE584 pKa = 4.14LEE586 pKa = 4.06QLVGSDD592 pKa = 4.07FQWPTSDD599 pKa = 3.05GWPSQEE605 pKa = 3.84FCLDD609 pKa = 3.53VYY611 pKa = 10.93LSPDD615 pKa = 3.25NKK617 pKa = 10.32IGGEE621 pKa = 4.11WYY623 pKa = 10.13SCVPSLEE630 pKa = 4.1EE631 pKa = 4.17DD632 pKa = 3.58LKK634 pKa = 11.07KK635 pKa = 10.71LRR637 pKa = 11.84SKK639 pKa = 10.61RR640 pKa = 11.84VTMEE644 pKa = 3.55AAKK647 pKa = 10.57LKK649 pKa = 9.75EE650 pKa = 4.05QGPKK654 pKa = 9.92FDD656 pKa = 3.94WALDD660 pKa = 3.46TDD662 pKa = 4.41LEE664 pKa = 4.52EE665 pKa = 5.04KK666 pKa = 10.59ASQLEE671 pKa = 3.91AVRR674 pKa = 11.84DD675 pKa = 3.98SKK677 pKa = 10.97IDD679 pKa = 3.4PRR681 pKa = 11.84VDD683 pKa = 3.1RR684 pKa = 11.84EE685 pKa = 4.3GYY687 pKa = 10.49LRR689 pKa = 11.84DD690 pKa = 3.62LVKK693 pKa = 10.42EE694 pKa = 4.15RR695 pKa = 11.84PLMRR699 pKa = 11.84PAKK702 pKa = 9.59QPKK705 pKa = 9.62NFLRR709 pKa = 11.84LKK711 pKa = 9.05GQEE714 pKa = 4.15SRR716 pKa = 11.84SKK718 pKa = 10.4DD719 pKa = 3.4QQIGDD724 pKa = 3.98FLEE727 pKa = 4.53KK728 pKa = 10.33FSSVLEE734 pKa = 4.03EE735 pKa = 5.44LPDD738 pKa = 3.48QMLEE742 pKa = 4.22LVVDD746 pKa = 4.55SPPGYY751 pKa = 9.85YY752 pKa = 10.02SRR754 pKa = 11.84VMQGKK759 pKa = 8.61GWRR762 pKa = 11.84PNPDD766 pKa = 2.6RR767 pKa = 11.84KK768 pKa = 9.69VWEE771 pKa = 4.5RR772 pKa = 11.84GSPVPVNPHH781 pKa = 3.23VWGRR785 pKa = 11.84DD786 pKa = 3.01ILMTLPDD793 pKa = 5.62FIPTEE798 pKa = 3.88KK799 pKa = 9.69KK800 pKa = 8.5TVRR803 pKa = 11.84KK804 pKa = 9.45EE805 pKa = 4.04CEE807 pKa = 3.61EE808 pKa = 4.4DD809 pKa = 3.81KK810 pKa = 10.05PTEE813 pKa = 4.19PFTLVPEE820 pKa = 4.3YY821 pKa = 11.19VKK823 pKa = 10.88LLPNVKK829 pKa = 8.89MDD831 pKa = 5.33AISLVTGSFSKK842 pKa = 10.99AGDD845 pKa = 3.16ILTTSNEE852 pKa = 4.11VLRR855 pKa = 11.84LSPDD859 pKa = 2.66SWIKK863 pKa = 9.66TEE865 pKa = 3.7VRR867 pKa = 11.84VRR869 pKa = 11.84NGTGIVARR877 pKa = 11.84AYY879 pKa = 10.31AKK881 pKa = 10.21SGKK884 pKa = 9.11AAKK887 pKa = 9.66EE888 pKa = 3.72MCFEE892 pKa = 4.56NMLEE896 pKa = 3.91QLKK899 pKa = 10.16EE900 pKa = 4.14RR901 pKa = 11.84EE902 pKa = 4.03RR903 pKa = 11.84QPTSFKK909 pKa = 10.78QQTEE913 pKa = 3.86EE914 pKa = 3.97DD915 pKa = 4.12GNRR918 pKa = 11.84STEE921 pKa = 4.0SEE923 pKa = 4.13AAAEE927 pKa = 4.28TQTTEE932 pKa = 4.03RR933 pKa = 11.84EE934 pKa = 4.19EE935 pKa = 4.28EE936 pKa = 3.95NGEE939 pKa = 4.16EE940 pKa = 4.19LVQDD944 pKa = 3.5VCEE947 pKa = 4.17VRR949 pKa = 11.84EE950 pKa = 4.32GTGGGDD956 pKa = 3.45GGVHH960 pKa = 6.6LL961 pKa = 5.16
Molecular weight: 109.3 kDa
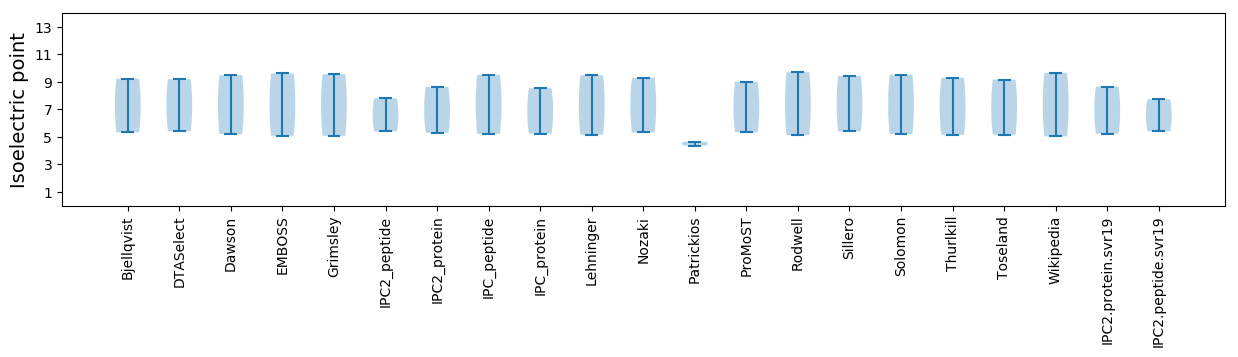
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHZ9|A0A1L3KHZ9_9VIRU RdRp OS=Hubei permutotetra-like virus 10 OX=1923074 PE=4 SV=1
MM1 pKa = 7.02ATEE4 pKa = 3.82AQKK7 pKa = 11.13AKK9 pKa = 10.45RR10 pKa = 11.84RR11 pKa = 11.84LKK13 pKa = 10.38RR14 pKa = 11.84KK15 pKa = 8.07RR16 pKa = 11.84QRR18 pKa = 11.84EE19 pKa = 4.07RR20 pKa = 11.84KK21 pKa = 9.69RR22 pKa = 11.84MEE24 pKa = 4.09KK25 pKa = 10.18NLSKK29 pKa = 10.51MSVKK33 pKa = 10.33SEE35 pKa = 3.8KK36 pKa = 10.63AQAAGTAGYY45 pKa = 9.66IFKK48 pKa = 9.42GTDD51 pKa = 2.94LVASVTVPQTAATGTIIARR70 pKa = 11.84LDD72 pKa = 3.14INPAKK77 pKa = 10.36FAGTTLAKK85 pKa = 10.13QASLWSNWIPRR96 pKa = 11.84KK97 pKa = 9.79LQVEE101 pKa = 4.42VQPSAGTTTSGTYY114 pKa = 10.19LVGWTLDD121 pKa = 3.44SALDD125 pKa = 3.58IPSGVNAIRR134 pKa = 11.84AVSAFEE140 pKa = 4.38KK141 pKa = 10.25PEE143 pKa = 3.78KK144 pKa = 10.11PRR146 pKa = 11.84IFDD149 pKa = 3.42KK150 pKa = 11.07VKK152 pKa = 10.78YY153 pKa = 10.1NISCKK158 pKa = 9.06MVQKK162 pKa = 10.27MLFCDD167 pKa = 4.91GSRR170 pKa = 11.84EE171 pKa = 4.25DD172 pKa = 3.99SDD174 pKa = 4.02QGTLYY179 pKa = 10.19IVLSSGIGNLVAGSVLTFNVYY200 pKa = 10.72LNYY203 pKa = 10.6DD204 pKa = 3.18IEE206 pKa = 4.83FYY208 pKa = 11.26NRR210 pKa = 11.84LATPASEE217 pKa = 4.27EE218 pKa = 4.02VLIYY222 pKa = 9.84PQAGYY227 pKa = 10.94EE228 pKa = 4.04NYY230 pKa = 8.49FTDD233 pKa = 5.36SSSDD237 pKa = 3.07WAGGTKK243 pKa = 10.48LSMKK247 pKa = 9.87QKK249 pKa = 10.41EE250 pKa = 4.46GGALVPWDD258 pKa = 3.65GAVPEE263 pKa = 5.39AIYY266 pKa = 10.25QFKK269 pKa = 9.03GTSLQYY275 pKa = 11.03YY276 pKa = 9.4DD277 pKa = 5.06AEE279 pKa = 4.51SALKK283 pKa = 10.08PITHH287 pKa = 7.46AVRR290 pKa = 11.84IRR292 pKa = 11.84DD293 pKa = 3.69RR294 pKa = 11.84GDD296 pKa = 2.84GALAVFEE303 pKa = 4.88SLAKK307 pKa = 9.62AQAYY311 pKa = 7.08ATTGDD316 pKa = 3.67EE317 pKa = 4.71VNCLTYY323 pKa = 10.33KK324 pKa = 10.02QAGPVVTPAGAPFMLVNPPPIFSNSLL350 pKa = 3.18
MM1 pKa = 7.02ATEE4 pKa = 3.82AQKK7 pKa = 11.13AKK9 pKa = 10.45RR10 pKa = 11.84RR11 pKa = 11.84LKK13 pKa = 10.38RR14 pKa = 11.84KK15 pKa = 8.07RR16 pKa = 11.84QRR18 pKa = 11.84EE19 pKa = 4.07RR20 pKa = 11.84KK21 pKa = 9.69RR22 pKa = 11.84MEE24 pKa = 4.09KK25 pKa = 10.18NLSKK29 pKa = 10.51MSVKK33 pKa = 10.33SEE35 pKa = 3.8KK36 pKa = 10.63AQAAGTAGYY45 pKa = 9.66IFKK48 pKa = 9.42GTDD51 pKa = 2.94LVASVTVPQTAATGTIIARR70 pKa = 11.84LDD72 pKa = 3.14INPAKK77 pKa = 10.36FAGTTLAKK85 pKa = 10.13QASLWSNWIPRR96 pKa = 11.84KK97 pKa = 9.79LQVEE101 pKa = 4.42VQPSAGTTTSGTYY114 pKa = 10.19LVGWTLDD121 pKa = 3.44SALDD125 pKa = 3.58IPSGVNAIRR134 pKa = 11.84AVSAFEE140 pKa = 4.38KK141 pKa = 10.25PEE143 pKa = 3.78KK144 pKa = 10.11PRR146 pKa = 11.84IFDD149 pKa = 3.42KK150 pKa = 11.07VKK152 pKa = 10.78YY153 pKa = 10.1NISCKK158 pKa = 9.06MVQKK162 pKa = 10.27MLFCDD167 pKa = 4.91GSRR170 pKa = 11.84EE171 pKa = 4.25DD172 pKa = 3.99SDD174 pKa = 4.02QGTLYY179 pKa = 10.19IVLSSGIGNLVAGSVLTFNVYY200 pKa = 10.72LNYY203 pKa = 10.6DD204 pKa = 3.18IEE206 pKa = 4.83FYY208 pKa = 11.26NRR210 pKa = 11.84LATPASEE217 pKa = 4.27EE218 pKa = 4.02VLIYY222 pKa = 9.84PQAGYY227 pKa = 10.94EE228 pKa = 4.04NYY230 pKa = 8.49FTDD233 pKa = 5.36SSSDD237 pKa = 3.07WAGGTKK243 pKa = 10.48LSMKK247 pKa = 9.87QKK249 pKa = 10.41EE250 pKa = 4.46GGALVPWDD258 pKa = 3.65GAVPEE263 pKa = 5.39AIYY266 pKa = 10.25QFKK269 pKa = 9.03GTSLQYY275 pKa = 11.03YY276 pKa = 9.4DD277 pKa = 5.06AEE279 pKa = 4.51SALKK283 pKa = 10.08PITHH287 pKa = 7.46AVRR290 pKa = 11.84IRR292 pKa = 11.84DD293 pKa = 3.69RR294 pKa = 11.84GDD296 pKa = 2.84GALAVFEE303 pKa = 4.88SLAKK307 pKa = 9.62AQAYY311 pKa = 7.08ATTGDD316 pKa = 3.67EE317 pKa = 4.71VNCLTYY323 pKa = 10.33KK324 pKa = 10.02QAGPVVTPAGAPFMLVNPPPIFSNSLL350 pKa = 3.18
Molecular weight: 38.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1311 |
350 |
961 |
655.5 |
73.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.865 ± 2.283 | 1.602 ± 0.373 |
5.492 ± 0.461 | 9.153 ± 2.149 |
3.966 ± 0.269 | 7.094 ± 0.167 |
0.686 ± 0.201 | 3.738 ± 0.56 |
8.085 ± 0.329 | 8.543 ± 0.415 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.983 ± 0.008 | 3.356 ± 0.179 |
5.492 ± 0.175 | 3.966 ± 0.16 |
5.645 ± 0.537 | 6.407 ± 0.654 |
5.645 ± 0.75 | 7.399 ± 0.271 |
1.754 ± 0.163 | 3.127 ± 0.58 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
