
Poecilia formosa (Amazon molly) (Limia formosa)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei;
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
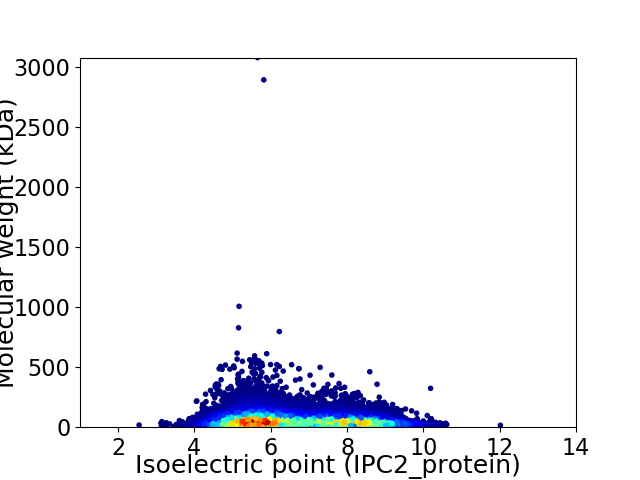
Virtual 2D-PAGE plot for 30440 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A087YD69|A0A087YD69_POEFO RAR related orphan receptor C OS=Poecilia formosa OX=48698 PE=3 SV=2
SS1 pKa = 7.36CPPFLQCQFNPCFPGVKK18 pKa = 9.57CVNTAPGFRR27 pKa = 11.84CDD29 pKa = 3.59ACPLGYY35 pKa = 10.54AGLPVEE41 pKa = 4.47GVGILFAQTNKK52 pKa = 9.79QVCDD56 pKa = 5.43DD57 pKa = 3.58IDD59 pKa = 3.69EE60 pKa = 4.9CKK62 pKa = 10.97GPDD65 pKa = 4.34NGGCTANSICHH76 pKa = 5.88NSVGSFYY83 pKa = 10.95CGSCKK88 pKa = 10.05TGFTGDD94 pKa = 3.44QVRR97 pKa = 11.84GCEE100 pKa = 4.19PEE102 pKa = 4.32LSCGNSLTNPCDD114 pKa = 3.35INAEE118 pKa = 4.42CIRR121 pKa = 11.84EE122 pKa = 3.91RR123 pKa = 11.84DD124 pKa = 3.67GSISCQCGIGWAGNGYY140 pKa = 9.95LCGKK144 pKa = 8.82DD145 pKa = 3.41TDD147 pKa = 3.57IDD149 pKa = 4.3GYY151 pKa = 10.17PDD153 pKa = 3.55EE154 pKa = 5.01KK155 pKa = 11.03LKK157 pKa = 11.4CKK159 pKa = 10.25DD160 pKa = 3.17ANCRR164 pKa = 11.84KK165 pKa = 9.73DD166 pKa = 3.26NCIFVPNSGQEE177 pKa = 3.85DD178 pKa = 4.13ADD180 pKa = 4.41RR181 pKa = 11.84DD182 pKa = 4.3GQGDD186 pKa = 3.88ACDD189 pKa = 4.9DD190 pKa = 4.28DD191 pKa = 6.95ADD193 pKa = 4.36GDD195 pKa = 4.52GIPNEE200 pKa = 4.16QDD202 pKa = 2.93NCRR205 pKa = 11.84LKK207 pKa = 11.13PNVDD211 pKa = 3.37QRR213 pKa = 11.84NSDD216 pKa = 3.65ADD218 pKa = 3.81SHH220 pKa = 7.54GDD222 pKa = 3.04ACDD225 pKa = 3.12NCRR228 pKa = 11.84VVNNPDD234 pKa = 3.29QRR236 pKa = 11.84DD237 pKa = 3.27TDD239 pKa = 3.86GDD241 pKa = 4.16GKK243 pKa = 11.42GDD245 pKa = 3.74ACDD248 pKa = 5.17DD249 pKa = 4.19DD250 pKa = 4.04MDD252 pKa = 6.26GDD254 pKa = 4.02GMIGVRR260 pKa = 11.84NFLDD264 pKa = 3.28NCQRR268 pKa = 11.84VQNRR272 pKa = 11.84DD273 pKa = 3.21QLDD276 pKa = 3.46RR277 pKa = 11.84DD278 pKa = 3.55GDD280 pKa = 4.42GVGDD284 pKa = 4.36ACDD287 pKa = 3.79SCPDD291 pKa = 3.6IPNPNQSDD299 pKa = 3.46VDD301 pKa = 3.63NDD303 pKa = 4.07LVGDD307 pKa = 4.11SCDD310 pKa = 3.63TNQDD314 pKa = 3.04SDD316 pKa = 4.1GDD318 pKa = 3.99GHH320 pKa = 7.29QDD322 pKa = 3.24TKK324 pKa = 11.65DD325 pKa = 3.4NCPLVINSSQLDD337 pKa = 3.61TDD339 pKa = 3.57KK340 pKa = 11.66DD341 pKa = 4.23GVGDD345 pKa = 4.08EE346 pKa = 5.46CDD348 pKa = 5.36DD349 pKa = 5.71DD350 pKa = 6.34DD351 pKa = 7.03DD352 pKa = 6.24NDD354 pKa = 5.44GIPDD358 pKa = 4.27TLPPGPDD365 pKa = 3.01NCRR368 pKa = 11.84LVPNPDD374 pKa = 3.79QIDD377 pKa = 3.91DD378 pKa = 4.08NNDD381 pKa = 3.08GVGDD385 pKa = 3.59ICEE388 pKa = 4.32SDD390 pKa = 3.29FDD392 pKa = 3.74QDD394 pKa = 4.05KK395 pKa = 11.41VIDD398 pKa = 5.09RR399 pKa = 11.84IDD401 pKa = 3.61NCPEE405 pKa = 3.68NAEE408 pKa = 4.12VTLTDD413 pKa = 3.81FRR415 pKa = 11.84AYY417 pKa = 8.19QTVVLDD423 pKa = 4.22PEE425 pKa = 5.32GDD427 pKa = 3.65AQIDD431 pKa = 4.13PNWVVLNQGMEE442 pKa = 4.11IVQTMNSDD450 pKa = 3.19PGLAVGYY457 pKa = 7.12TAFSGVDD464 pKa = 3.72FEE466 pKa = 5.24GTFHH470 pKa = 6.81VNTVTDD476 pKa = 3.57DD477 pKa = 4.08DD478 pKa = 4.35YY479 pKa = 12.0AGFIFGYY486 pKa = 9.58QDD488 pKa = 2.83SSSFYY493 pKa = 10.47VVMWKK498 pKa = 7.78QTEE501 pKa = 3.86QTYY504 pKa = 8.13WQATPFRR511 pKa = 11.84AVAEE515 pKa = 4.17PGIQLKK521 pKa = 10.07AVKK524 pKa = 9.92SRR526 pKa = 11.84SGPGEE531 pKa = 3.8HH532 pKa = 6.66LRR534 pKa = 11.84NSLWHH539 pKa = 6.47TGDD542 pKa = 3.39TNDD545 pKa = 4.93QVRR548 pKa = 11.84LLWKK552 pKa = 10.3DD553 pKa = 3.22PRR555 pKa = 11.84NVGWKK560 pKa = 10.39DD561 pKa = 3.16KK562 pKa = 11.21VSYY565 pKa = 10.03RR566 pKa = 11.84WYY568 pKa = 9.83LQHH571 pKa = 7.1RR572 pKa = 11.84PQIGYY577 pKa = 9.15IRR579 pKa = 11.84VRR581 pKa = 11.84FYY583 pKa = 11.37EE584 pKa = 4.3GTQLVADD591 pKa = 4.13SGVTIDD597 pKa = 3.09TTMRR601 pKa = 11.84GGRR604 pKa = 11.84LGVFCFSQEE613 pKa = 3.94NIIWSNLKK621 pKa = 9.68YY622 pKa = 10.31RR623 pKa = 11.84CNDD626 pKa = 4.18TIPEE630 pKa = 4.24DD631 pKa = 3.8FQEE634 pKa = 5.2FSAQHH639 pKa = 5.35TVDD642 pKa = 4.21SII644 pKa = 3.74
SS1 pKa = 7.36CPPFLQCQFNPCFPGVKK18 pKa = 9.57CVNTAPGFRR27 pKa = 11.84CDD29 pKa = 3.59ACPLGYY35 pKa = 10.54AGLPVEE41 pKa = 4.47GVGILFAQTNKK52 pKa = 9.79QVCDD56 pKa = 5.43DD57 pKa = 3.58IDD59 pKa = 3.69EE60 pKa = 4.9CKK62 pKa = 10.97GPDD65 pKa = 4.34NGGCTANSICHH76 pKa = 5.88NSVGSFYY83 pKa = 10.95CGSCKK88 pKa = 10.05TGFTGDD94 pKa = 3.44QVRR97 pKa = 11.84GCEE100 pKa = 4.19PEE102 pKa = 4.32LSCGNSLTNPCDD114 pKa = 3.35INAEE118 pKa = 4.42CIRR121 pKa = 11.84EE122 pKa = 3.91RR123 pKa = 11.84DD124 pKa = 3.67GSISCQCGIGWAGNGYY140 pKa = 9.95LCGKK144 pKa = 8.82DD145 pKa = 3.41TDD147 pKa = 3.57IDD149 pKa = 4.3GYY151 pKa = 10.17PDD153 pKa = 3.55EE154 pKa = 5.01KK155 pKa = 11.03LKK157 pKa = 11.4CKK159 pKa = 10.25DD160 pKa = 3.17ANCRR164 pKa = 11.84KK165 pKa = 9.73DD166 pKa = 3.26NCIFVPNSGQEE177 pKa = 3.85DD178 pKa = 4.13ADD180 pKa = 4.41RR181 pKa = 11.84DD182 pKa = 4.3GQGDD186 pKa = 3.88ACDD189 pKa = 4.9DD190 pKa = 4.28DD191 pKa = 6.95ADD193 pKa = 4.36GDD195 pKa = 4.52GIPNEE200 pKa = 4.16QDD202 pKa = 2.93NCRR205 pKa = 11.84LKK207 pKa = 11.13PNVDD211 pKa = 3.37QRR213 pKa = 11.84NSDD216 pKa = 3.65ADD218 pKa = 3.81SHH220 pKa = 7.54GDD222 pKa = 3.04ACDD225 pKa = 3.12NCRR228 pKa = 11.84VVNNPDD234 pKa = 3.29QRR236 pKa = 11.84DD237 pKa = 3.27TDD239 pKa = 3.86GDD241 pKa = 4.16GKK243 pKa = 11.42GDD245 pKa = 3.74ACDD248 pKa = 5.17DD249 pKa = 4.19DD250 pKa = 4.04MDD252 pKa = 6.26GDD254 pKa = 4.02GMIGVRR260 pKa = 11.84NFLDD264 pKa = 3.28NCQRR268 pKa = 11.84VQNRR272 pKa = 11.84DD273 pKa = 3.21QLDD276 pKa = 3.46RR277 pKa = 11.84DD278 pKa = 3.55GDD280 pKa = 4.42GVGDD284 pKa = 4.36ACDD287 pKa = 3.79SCPDD291 pKa = 3.6IPNPNQSDD299 pKa = 3.46VDD301 pKa = 3.63NDD303 pKa = 4.07LVGDD307 pKa = 4.11SCDD310 pKa = 3.63TNQDD314 pKa = 3.04SDD316 pKa = 4.1GDD318 pKa = 3.99GHH320 pKa = 7.29QDD322 pKa = 3.24TKK324 pKa = 11.65DD325 pKa = 3.4NCPLVINSSQLDD337 pKa = 3.61TDD339 pKa = 3.57KK340 pKa = 11.66DD341 pKa = 4.23GVGDD345 pKa = 4.08EE346 pKa = 5.46CDD348 pKa = 5.36DD349 pKa = 5.71DD350 pKa = 6.34DD351 pKa = 7.03DD352 pKa = 6.24NDD354 pKa = 5.44GIPDD358 pKa = 4.27TLPPGPDD365 pKa = 3.01NCRR368 pKa = 11.84LVPNPDD374 pKa = 3.79QIDD377 pKa = 3.91DD378 pKa = 4.08NNDD381 pKa = 3.08GVGDD385 pKa = 3.59ICEE388 pKa = 4.32SDD390 pKa = 3.29FDD392 pKa = 3.74QDD394 pKa = 4.05KK395 pKa = 11.41VIDD398 pKa = 5.09RR399 pKa = 11.84IDD401 pKa = 3.61NCPEE405 pKa = 3.68NAEE408 pKa = 4.12VTLTDD413 pKa = 3.81FRR415 pKa = 11.84AYY417 pKa = 8.19QTVVLDD423 pKa = 4.22PEE425 pKa = 5.32GDD427 pKa = 3.65AQIDD431 pKa = 4.13PNWVVLNQGMEE442 pKa = 4.11IVQTMNSDD450 pKa = 3.19PGLAVGYY457 pKa = 7.12TAFSGVDD464 pKa = 3.72FEE466 pKa = 5.24GTFHH470 pKa = 6.81VNTVTDD476 pKa = 3.57DD477 pKa = 4.08DD478 pKa = 4.35YY479 pKa = 12.0AGFIFGYY486 pKa = 9.58QDD488 pKa = 2.83SSSFYY493 pKa = 10.47VVMWKK498 pKa = 7.78QTEE501 pKa = 3.86QTYY504 pKa = 8.13WQATPFRR511 pKa = 11.84AVAEE515 pKa = 4.17PGIQLKK521 pKa = 10.07AVKK524 pKa = 9.92SRR526 pKa = 11.84SGPGEE531 pKa = 3.8HH532 pKa = 6.66LRR534 pKa = 11.84NSLWHH539 pKa = 6.47TGDD542 pKa = 3.39TNDD545 pKa = 4.93QVRR548 pKa = 11.84LLWKK552 pKa = 10.3DD553 pKa = 3.22PRR555 pKa = 11.84NVGWKK560 pKa = 10.39DD561 pKa = 3.16KK562 pKa = 11.21VSYY565 pKa = 10.03RR566 pKa = 11.84WYY568 pKa = 9.83LQHH571 pKa = 7.1RR572 pKa = 11.84PQIGYY577 pKa = 9.15IRR579 pKa = 11.84VRR581 pKa = 11.84FYY583 pKa = 11.37EE584 pKa = 4.3GTQLVADD591 pKa = 4.13SGVTIDD597 pKa = 3.09TTMRR601 pKa = 11.84GGRR604 pKa = 11.84LGVFCFSQEE613 pKa = 3.94NIIWSNLKK621 pKa = 9.68YY622 pKa = 10.31RR623 pKa = 11.84CNDD626 pKa = 4.18TIPEE630 pKa = 4.24DD631 pKa = 3.8FQEE634 pKa = 5.2FSAQHH639 pKa = 5.35TVDD642 pKa = 4.21SII644 pKa = 3.74
Molecular weight: 70.52 kDa
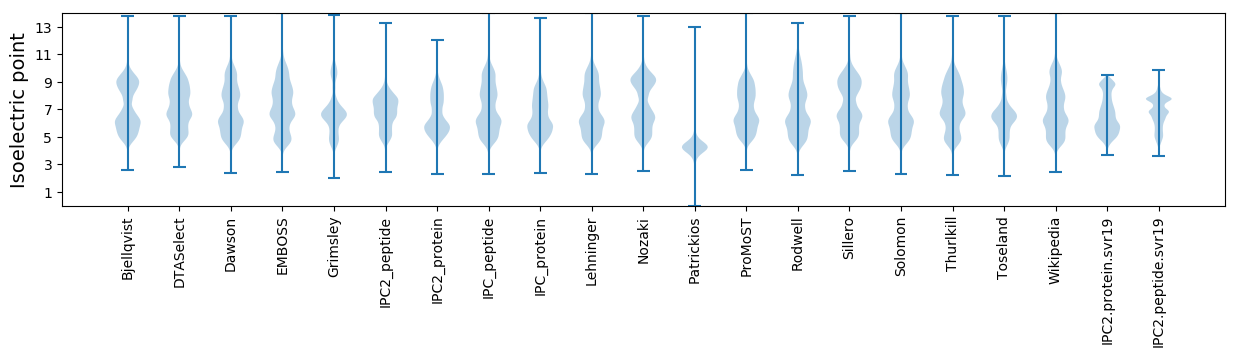
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A087YRC7|A0A087YRC7_POEFO 5-hydroxytryptamine receptor 1B OS=Poecilia formosa OX=48698 GN=HTR1B PE=3 SV=2
GG1 pKa = 7.23GGPGRR6 pKa = 11.84PKK8 pKa = 10.4SGGPGRR14 pKa = 11.84PKK16 pKa = 10.24PGGGGGPGRR25 pKa = 11.84PKK27 pKa = 10.19PGGPGRR33 pKa = 11.84PKK35 pKa = 10.2PGGGGGPGRR44 pKa = 11.84PKK46 pKa = 10.3PGGGGGPGRR55 pKa = 11.84PKK57 pKa = 10.3PGGGGGPGRR66 pKa = 11.84PTT68 pKa = 3.21
GG1 pKa = 7.23GGPGRR6 pKa = 11.84PKK8 pKa = 10.4SGGPGRR14 pKa = 11.84PKK16 pKa = 10.24PGGGGGPGRR25 pKa = 11.84PKK27 pKa = 10.19PGGPGRR33 pKa = 11.84PKK35 pKa = 10.2PGGGGGPGRR44 pKa = 11.84PKK46 pKa = 10.3PGGGGGPGRR55 pKa = 11.84PKK57 pKa = 10.3PGGGGGPGRR66 pKa = 11.84PTT68 pKa = 3.21
Molecular weight: 5.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
17431430 |
27 |
27691 |
572.6 |
63.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.527 ± 0.012 | 2.295 ± 0.012 |
5.188 ± 0.009 | 6.868 ± 0.019 |
3.684 ± 0.01 | 6.279 ± 0.016 |
2.609 ± 0.008 | 4.328 ± 0.012 |
5.758 ± 0.017 | 9.552 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.006 | 3.877 ± 0.008 |
5.714 ± 0.02 | 4.754 ± 0.014 |
5.681 ± 0.012 | 8.903 ± 0.02 |
5.562 ± 0.011 | 6.301 ± 0.016 |
1.148 ± 0.004 | 2.674 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
