
Fenneropenaeus chinensis hepatopancreatic densovirus (isolate Shrimp/China/HB/2008)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Densovirinae; Hepandensovirus; Decapod hepandensovirus 1; Fenneropenaeus chinensis hepandensovirus
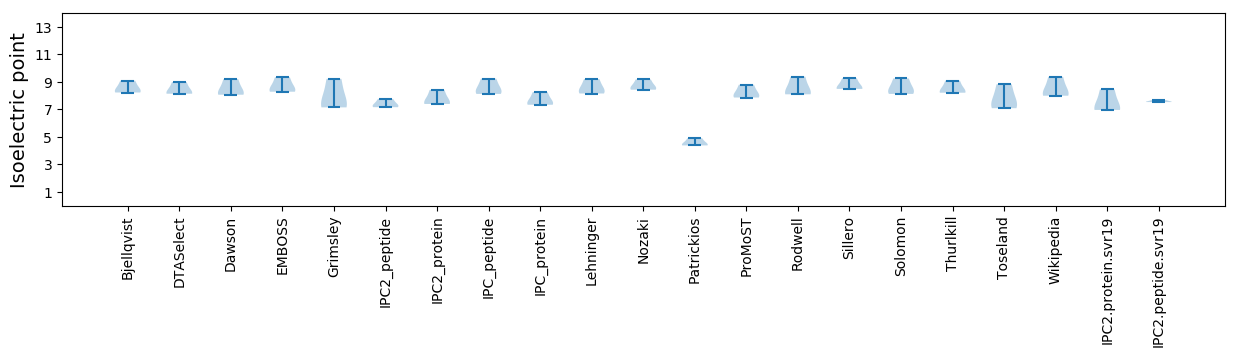
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
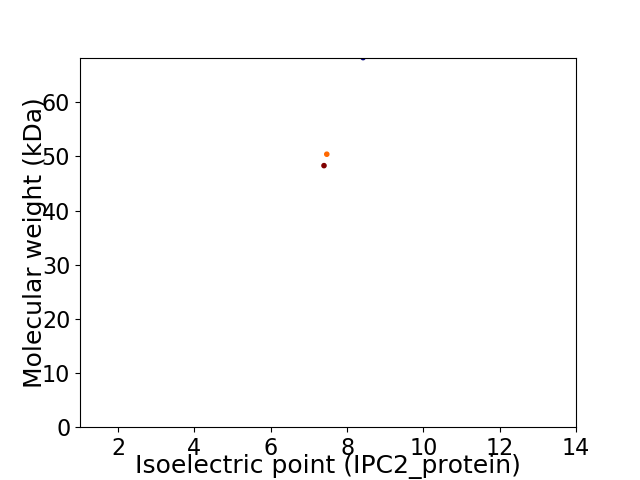
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E2E2D1|E2E2D1_FCDNV Truncated structural protein OS=Fenneropenaeus chinensis hepatopancreatic densovirus (isolate Shrimp/China/HB/2008) OX=1556324 GN=CP PE=4 SV=1
MM1 pKa = 7.54SPTRR5 pKa = 11.84KK6 pKa = 9.67GGNYY10 pKa = 8.53FASKK14 pKa = 10.31HH15 pKa = 4.7FQSKK19 pKa = 10.11KK20 pKa = 9.13KK21 pKa = 10.55NKK23 pKa = 9.3LGKK26 pKa = 10.6VKK28 pKa = 10.68DD29 pKa = 3.75LLASKK34 pKa = 10.43KK35 pKa = 10.17KK36 pKa = 9.24EE37 pKa = 3.79RR38 pKa = 11.84KK39 pKa = 9.33FKK41 pKa = 11.23GKK43 pKa = 10.76GNTLSEE49 pKa = 4.43EE50 pKa = 4.36PSTSGWKK57 pKa = 10.55DD58 pKa = 3.24PVRR61 pKa = 11.84QRR63 pKa = 11.84FPALEE68 pKa = 3.86QEE70 pKa = 4.41EE71 pKa = 4.88RR72 pKa = 11.84NTFAGLLAIEE82 pKa = 4.84AAPDD86 pKa = 3.36QRR88 pKa = 11.84QLGRR92 pKa = 11.84DD93 pKa = 3.64SNNQLALVQRR103 pKa = 11.84DD104 pKa = 3.21TRR106 pKa = 11.84VAVRR110 pKa = 11.84QSTNRR115 pKa = 11.84RR116 pKa = 11.84EE117 pKa = 3.98ALEE120 pKa = 4.16VVRR123 pKa = 11.84AATQAIRR130 pKa = 11.84SGGDD134 pKa = 2.78RR135 pKa = 11.84LAEE138 pKa = 3.91LVQTYY143 pKa = 11.05ASGFSDD149 pKa = 3.04STEE152 pKa = 3.73IVEE155 pKa = 5.12LRR157 pKa = 11.84QEE159 pKa = 4.14DD160 pKa = 4.44RR161 pKa = 11.84IQRR164 pKa = 11.84DD165 pKa = 2.98IFQEE169 pKa = 3.97EE170 pKa = 4.76GQNLLAIEE178 pKa = 4.33IALEE182 pKa = 4.68GPSSVTQQFDD192 pKa = 3.5QEE194 pKa = 4.07KK195 pKa = 8.17TPAVKK200 pKa = 10.14RR201 pKa = 11.84ALEE204 pKa = 3.98LTQEE208 pKa = 4.21EE209 pKa = 4.48EE210 pKa = 3.72QLEE213 pKa = 4.38RR214 pKa = 11.84IEE216 pKa = 4.09NAKK219 pKa = 10.42KK220 pKa = 10.62YY221 pKa = 9.14IEE223 pKa = 4.1EE224 pKa = 4.37VIEE227 pKa = 3.86EE228 pKa = 4.31TNRR231 pKa = 11.84EE232 pKa = 3.79FEE234 pKa = 4.2DD235 pKa = 3.5QVRR238 pKa = 11.84QEE240 pKa = 4.25TSASAEE246 pKa = 3.95DD247 pKa = 4.12TMASAAPTPMEE258 pKa = 3.89TSEE261 pKa = 4.57PGVTAAPHH269 pKa = 5.26QKK271 pKa = 9.87SAAGGGGGGGSGGEE285 pKa = 3.79TAGYY289 pKa = 9.99GKK291 pKa = 8.49NTNDD295 pKa = 2.63AFQRR299 pKa = 11.84HH300 pKa = 5.5RR301 pKa = 11.84NRR303 pKa = 11.84PIDD306 pKa = 3.9LKK308 pKa = 11.21HH309 pKa = 7.3IGDD312 pKa = 4.01NVYY315 pKa = 10.11VAQRR319 pKa = 11.84VYY321 pKa = 10.73KK322 pKa = 10.8VEE324 pKa = 4.25AEE326 pKa = 4.33CKK328 pKa = 10.71NIDD331 pKa = 5.09EE332 pKa = 4.64KK333 pKa = 11.16LTWSNTANNEE343 pKa = 3.75YY344 pKa = 9.68TRR346 pKa = 11.84RR347 pKa = 11.84LMGTNGNSDD356 pKa = 3.27KK357 pKa = 11.66GNVKK361 pKa = 10.37FNFTAPINGSIGMGNLALGNYY382 pKa = 8.01INAWGIDD389 pKa = 3.87NIAKK393 pKa = 10.25SEE395 pKa = 4.13DD396 pKa = 2.99SWAIIATRR404 pKa = 11.84GKK406 pKa = 9.59MNHH409 pKa = 5.66LQAFEE414 pKa = 4.58MIPPNARR421 pKa = 11.84RR422 pKa = 11.84NCSRR426 pKa = 11.84VYY428 pKa = 10.3KK429 pKa = 10.44CSSTVWW435 pKa = 3.08
MM1 pKa = 7.54SPTRR5 pKa = 11.84KK6 pKa = 9.67GGNYY10 pKa = 8.53FASKK14 pKa = 10.31HH15 pKa = 4.7FQSKK19 pKa = 10.11KK20 pKa = 9.13KK21 pKa = 10.55NKK23 pKa = 9.3LGKK26 pKa = 10.6VKK28 pKa = 10.68DD29 pKa = 3.75LLASKK34 pKa = 10.43KK35 pKa = 10.17KK36 pKa = 9.24EE37 pKa = 3.79RR38 pKa = 11.84KK39 pKa = 9.33FKK41 pKa = 11.23GKK43 pKa = 10.76GNTLSEE49 pKa = 4.43EE50 pKa = 4.36PSTSGWKK57 pKa = 10.55DD58 pKa = 3.24PVRR61 pKa = 11.84QRR63 pKa = 11.84FPALEE68 pKa = 3.86QEE70 pKa = 4.41EE71 pKa = 4.88RR72 pKa = 11.84NTFAGLLAIEE82 pKa = 4.84AAPDD86 pKa = 3.36QRR88 pKa = 11.84QLGRR92 pKa = 11.84DD93 pKa = 3.64SNNQLALVQRR103 pKa = 11.84DD104 pKa = 3.21TRR106 pKa = 11.84VAVRR110 pKa = 11.84QSTNRR115 pKa = 11.84RR116 pKa = 11.84EE117 pKa = 3.98ALEE120 pKa = 4.16VVRR123 pKa = 11.84AATQAIRR130 pKa = 11.84SGGDD134 pKa = 2.78RR135 pKa = 11.84LAEE138 pKa = 3.91LVQTYY143 pKa = 11.05ASGFSDD149 pKa = 3.04STEE152 pKa = 3.73IVEE155 pKa = 5.12LRR157 pKa = 11.84QEE159 pKa = 4.14DD160 pKa = 4.44RR161 pKa = 11.84IQRR164 pKa = 11.84DD165 pKa = 2.98IFQEE169 pKa = 3.97EE170 pKa = 4.76GQNLLAIEE178 pKa = 4.33IALEE182 pKa = 4.68GPSSVTQQFDD192 pKa = 3.5QEE194 pKa = 4.07KK195 pKa = 8.17TPAVKK200 pKa = 10.14RR201 pKa = 11.84ALEE204 pKa = 3.98LTQEE208 pKa = 4.21EE209 pKa = 4.48EE210 pKa = 3.72QLEE213 pKa = 4.38RR214 pKa = 11.84IEE216 pKa = 4.09NAKK219 pKa = 10.42KK220 pKa = 10.62YY221 pKa = 9.14IEE223 pKa = 4.1EE224 pKa = 4.37VIEE227 pKa = 3.86EE228 pKa = 4.31TNRR231 pKa = 11.84EE232 pKa = 3.79FEE234 pKa = 4.2DD235 pKa = 3.5QVRR238 pKa = 11.84QEE240 pKa = 4.25TSASAEE246 pKa = 3.95DD247 pKa = 4.12TMASAAPTPMEE258 pKa = 3.89TSEE261 pKa = 4.57PGVTAAPHH269 pKa = 5.26QKK271 pKa = 9.87SAAGGGGGGGSGGEE285 pKa = 3.79TAGYY289 pKa = 9.99GKK291 pKa = 8.49NTNDD295 pKa = 2.63AFQRR299 pKa = 11.84HH300 pKa = 5.5RR301 pKa = 11.84NRR303 pKa = 11.84PIDD306 pKa = 3.9LKK308 pKa = 11.21HH309 pKa = 7.3IGDD312 pKa = 4.01NVYY315 pKa = 10.11VAQRR319 pKa = 11.84VYY321 pKa = 10.73KK322 pKa = 10.8VEE324 pKa = 4.25AEE326 pKa = 4.33CKK328 pKa = 10.71NIDD331 pKa = 5.09EE332 pKa = 4.64KK333 pKa = 11.16LTWSNTANNEE343 pKa = 3.75YY344 pKa = 9.68TRR346 pKa = 11.84RR347 pKa = 11.84LMGTNGNSDD356 pKa = 3.27KK357 pKa = 11.66GNVKK361 pKa = 10.37FNFTAPINGSIGMGNLALGNYY382 pKa = 8.01INAWGIDD389 pKa = 3.87NIAKK393 pKa = 10.25SEE395 pKa = 4.13DD396 pKa = 2.99SWAIIATRR404 pKa = 11.84GKK406 pKa = 9.59MNHH409 pKa = 5.66LQAFEE414 pKa = 4.58MIPPNARR421 pKa = 11.84RR422 pKa = 11.84NCSRR426 pKa = 11.84VYY428 pKa = 10.3KK429 pKa = 10.44CSSTVWW435 pKa = 3.08
Molecular weight: 48.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E2E2D1|E2E2D1_FCDNV Truncated structural protein OS=Fenneropenaeus chinensis hepatopancreatic densovirus (isolate Shrimp/China/HB/2008) OX=1556324 GN=CP PE=4 SV=1
MM1 pKa = 7.14FRR3 pKa = 11.84QVMKK7 pKa = 10.6LKK9 pKa = 10.53RR10 pKa = 11.84EE11 pKa = 3.99GMVDD15 pKa = 3.56HH16 pKa = 6.85NPLVTFYY23 pKa = 11.15SGIIVKK29 pKa = 10.17FEE31 pKa = 3.69HH32 pKa = 6.45WNNNVSKK39 pKa = 10.65VRR41 pKa = 11.84KK42 pKa = 8.9FLYY45 pKa = 10.37KK46 pKa = 10.25FAQWLYY52 pKa = 10.68KK53 pKa = 9.93EE54 pKa = 4.31CTYY57 pKa = 10.79IHH59 pKa = 6.67NISAAVHH66 pKa = 6.28DD67 pKa = 4.36RR68 pKa = 11.84CKK70 pKa = 10.97DD71 pKa = 3.59DD72 pKa = 3.71CCKK75 pKa = 11.01DD76 pKa = 3.48SANKK80 pKa = 9.09VCKK83 pKa = 10.34NIYY86 pKa = 9.71GPHH89 pKa = 5.59LHH91 pKa = 6.93ILLEE95 pKa = 4.46SVNEE99 pKa = 4.01NWSKK103 pKa = 10.24SSKK106 pKa = 9.43RR107 pKa = 11.84VLFRR111 pKa = 11.84GYY113 pKa = 10.66EE114 pKa = 3.98KK115 pKa = 10.38ILQHH119 pKa = 6.0EE120 pKa = 4.48NKK122 pKa = 9.96QLWEE126 pKa = 4.18DD127 pKa = 3.62LGLQKK132 pKa = 9.75TSPPSMSLWDD142 pKa = 3.46GEE144 pKa = 4.3MFKK147 pKa = 10.43WYY149 pKa = 9.05MFRR152 pKa = 11.84DD153 pKa = 3.22RR154 pKa = 11.84KK155 pKa = 8.23YY156 pKa = 9.98AKK158 pKa = 10.04VHH160 pKa = 4.11GTYY163 pKa = 10.15YY164 pKa = 10.34YY165 pKa = 11.0SSNEE169 pKa = 3.77EE170 pKa = 4.16LLSKK174 pKa = 9.86LVKK177 pKa = 10.36MKK179 pKa = 9.44DD180 pKa = 3.14TQEE183 pKa = 4.66RR184 pKa = 11.84DD185 pKa = 3.3DD186 pKa = 5.25LYY188 pKa = 11.59EE189 pKa = 4.19KK190 pKa = 10.82ACQFKK195 pKa = 10.39KK196 pKa = 10.78DD197 pKa = 3.26KK198 pKa = 10.64NAARR202 pKa = 11.84KK203 pKa = 9.47IEE205 pKa = 4.08NSTAKK210 pKa = 9.69TLDD213 pKa = 3.61GADD216 pKa = 3.48SDD218 pKa = 4.58NIRR221 pKa = 11.84LSSSRR226 pKa = 11.84AIYY229 pKa = 10.52LEE231 pKa = 3.77NLQVLEE237 pKa = 4.51KK238 pKa = 10.97YY239 pKa = 9.74LVKK242 pKa = 10.5HH243 pKa = 5.53KK244 pKa = 10.49CYY246 pKa = 10.12TIQDD250 pKa = 3.9FKK252 pKa = 11.29MMQRR256 pKa = 11.84NEE258 pKa = 3.77DD259 pKa = 4.41EE260 pKa = 3.64IWVNYY265 pKa = 9.59MYY267 pKa = 10.34DD268 pKa = 3.18IQNLEE273 pKa = 3.97KK274 pKa = 10.85VIEE277 pKa = 4.25KK278 pKa = 10.44LNIMEE283 pKa = 4.71YY284 pKa = 10.69SLQQADD290 pKa = 4.32YY291 pKa = 10.9IEE293 pKa = 4.88GNTWIGEE300 pKa = 4.27DD301 pKa = 3.24LWNTNSAYY309 pKa = 9.59MRR311 pKa = 11.84TVKK314 pKa = 10.47RR315 pKa = 11.84GTDD318 pKa = 2.6RR319 pKa = 11.84YY320 pKa = 8.05YY321 pKa = 10.05WYY323 pKa = 9.9IQRR326 pKa = 11.84HH327 pKa = 4.17VSNRR331 pKa = 11.84ASLIGQSRR339 pKa = 11.84QICIDD344 pKa = 3.29GAYY347 pKa = 10.15MMFNIIDD354 pKa = 3.64NMRR357 pKa = 11.84VEE359 pKa = 4.74SRR361 pKa = 11.84PKK363 pKa = 9.43TIPIISKK370 pKa = 9.94NKK372 pKa = 6.87TVQWIQDD379 pKa = 3.46FMDD382 pKa = 5.22IIHH385 pKa = 6.93GNLPKK390 pKa = 10.04INCMMLYY397 pKa = 10.73GNSNSGKK404 pKa = 7.27TQLIEE409 pKa = 4.17ALTGLVNTAIMTNVGDD425 pKa = 3.57GGTFHH430 pKa = 7.44FSNITEE436 pKa = 3.89MSTIVVGNEE445 pKa = 3.64TKK447 pKa = 10.34IRR449 pKa = 11.84TQTIEE454 pKa = 3.66QWKK457 pKa = 8.9GLCGGEE463 pKa = 3.99NVTMPMKK470 pKa = 10.34YY471 pKa = 9.43KK472 pKa = 9.73EE473 pKa = 4.68HH474 pKa = 5.97KK475 pKa = 7.9THH477 pKa = 6.63MFRR480 pKa = 11.84KK481 pKa = 9.29PVFLTNQHH489 pKa = 6.26HH490 pKa = 6.51PLVDD494 pKa = 3.13ISHH497 pKa = 6.45YY498 pKa = 10.31DD499 pKa = 3.36DD500 pKa = 4.04RR501 pKa = 11.84RR502 pKa = 11.84AIEE505 pKa = 4.28NRR507 pKa = 11.84SFMYY511 pKa = 10.08KK512 pKa = 9.92VEE514 pKa = 4.53LGSEE518 pKa = 4.2PVNAHH523 pKa = 5.48IKK525 pKa = 10.03FPNRR529 pKa = 11.84MIPIKK534 pKa = 10.4KK535 pKa = 9.95NPEE538 pKa = 3.49LTQFVLASMQYY549 pKa = 9.65VHH551 pKa = 6.69TNYY554 pKa = 9.66MNRR557 pKa = 11.84PDD559 pKa = 3.86KK560 pKa = 10.74KK561 pKa = 10.69FKK563 pKa = 10.42IGFFNKK569 pKa = 10.31LYY571 pKa = 10.73DD572 pKa = 3.86ALFEE576 pKa = 4.33NSS578 pKa = 3.11
MM1 pKa = 7.14FRR3 pKa = 11.84QVMKK7 pKa = 10.6LKK9 pKa = 10.53RR10 pKa = 11.84EE11 pKa = 3.99GMVDD15 pKa = 3.56HH16 pKa = 6.85NPLVTFYY23 pKa = 11.15SGIIVKK29 pKa = 10.17FEE31 pKa = 3.69HH32 pKa = 6.45WNNNVSKK39 pKa = 10.65VRR41 pKa = 11.84KK42 pKa = 8.9FLYY45 pKa = 10.37KK46 pKa = 10.25FAQWLYY52 pKa = 10.68KK53 pKa = 9.93EE54 pKa = 4.31CTYY57 pKa = 10.79IHH59 pKa = 6.67NISAAVHH66 pKa = 6.28DD67 pKa = 4.36RR68 pKa = 11.84CKK70 pKa = 10.97DD71 pKa = 3.59DD72 pKa = 3.71CCKK75 pKa = 11.01DD76 pKa = 3.48SANKK80 pKa = 9.09VCKK83 pKa = 10.34NIYY86 pKa = 9.71GPHH89 pKa = 5.59LHH91 pKa = 6.93ILLEE95 pKa = 4.46SVNEE99 pKa = 4.01NWSKK103 pKa = 10.24SSKK106 pKa = 9.43RR107 pKa = 11.84VLFRR111 pKa = 11.84GYY113 pKa = 10.66EE114 pKa = 3.98KK115 pKa = 10.38ILQHH119 pKa = 6.0EE120 pKa = 4.48NKK122 pKa = 9.96QLWEE126 pKa = 4.18DD127 pKa = 3.62LGLQKK132 pKa = 9.75TSPPSMSLWDD142 pKa = 3.46GEE144 pKa = 4.3MFKK147 pKa = 10.43WYY149 pKa = 9.05MFRR152 pKa = 11.84DD153 pKa = 3.22RR154 pKa = 11.84KK155 pKa = 8.23YY156 pKa = 9.98AKK158 pKa = 10.04VHH160 pKa = 4.11GTYY163 pKa = 10.15YY164 pKa = 10.34YY165 pKa = 11.0SSNEE169 pKa = 3.77EE170 pKa = 4.16LLSKK174 pKa = 9.86LVKK177 pKa = 10.36MKK179 pKa = 9.44DD180 pKa = 3.14TQEE183 pKa = 4.66RR184 pKa = 11.84DD185 pKa = 3.3DD186 pKa = 5.25LYY188 pKa = 11.59EE189 pKa = 4.19KK190 pKa = 10.82ACQFKK195 pKa = 10.39KK196 pKa = 10.78DD197 pKa = 3.26KK198 pKa = 10.64NAARR202 pKa = 11.84KK203 pKa = 9.47IEE205 pKa = 4.08NSTAKK210 pKa = 9.69TLDD213 pKa = 3.61GADD216 pKa = 3.48SDD218 pKa = 4.58NIRR221 pKa = 11.84LSSSRR226 pKa = 11.84AIYY229 pKa = 10.52LEE231 pKa = 3.77NLQVLEE237 pKa = 4.51KK238 pKa = 10.97YY239 pKa = 9.74LVKK242 pKa = 10.5HH243 pKa = 5.53KK244 pKa = 10.49CYY246 pKa = 10.12TIQDD250 pKa = 3.9FKK252 pKa = 11.29MMQRR256 pKa = 11.84NEE258 pKa = 3.77DD259 pKa = 4.41EE260 pKa = 3.64IWVNYY265 pKa = 9.59MYY267 pKa = 10.34DD268 pKa = 3.18IQNLEE273 pKa = 3.97KK274 pKa = 10.85VIEE277 pKa = 4.25KK278 pKa = 10.44LNIMEE283 pKa = 4.71YY284 pKa = 10.69SLQQADD290 pKa = 4.32YY291 pKa = 10.9IEE293 pKa = 4.88GNTWIGEE300 pKa = 4.27DD301 pKa = 3.24LWNTNSAYY309 pKa = 9.59MRR311 pKa = 11.84TVKK314 pKa = 10.47RR315 pKa = 11.84GTDD318 pKa = 2.6RR319 pKa = 11.84YY320 pKa = 8.05YY321 pKa = 10.05WYY323 pKa = 9.9IQRR326 pKa = 11.84HH327 pKa = 4.17VSNRR331 pKa = 11.84ASLIGQSRR339 pKa = 11.84QICIDD344 pKa = 3.29GAYY347 pKa = 10.15MMFNIIDD354 pKa = 3.64NMRR357 pKa = 11.84VEE359 pKa = 4.74SRR361 pKa = 11.84PKK363 pKa = 9.43TIPIISKK370 pKa = 9.94NKK372 pKa = 6.87TVQWIQDD379 pKa = 3.46FMDD382 pKa = 5.22IIHH385 pKa = 6.93GNLPKK390 pKa = 10.04INCMMLYY397 pKa = 10.73GNSNSGKK404 pKa = 7.27TQLIEE409 pKa = 4.17ALTGLVNTAIMTNVGDD425 pKa = 3.57GGTFHH430 pKa = 7.44FSNITEE436 pKa = 3.89MSTIVVGNEE445 pKa = 3.64TKK447 pKa = 10.34IRR449 pKa = 11.84TQTIEE454 pKa = 3.66QWKK457 pKa = 8.9GLCGGEE463 pKa = 3.99NVTMPMKK470 pKa = 10.34YY471 pKa = 9.43KK472 pKa = 9.73EE473 pKa = 4.68HH474 pKa = 5.97KK475 pKa = 7.9THH477 pKa = 6.63MFRR480 pKa = 11.84KK481 pKa = 9.29PVFLTNQHH489 pKa = 6.26HH490 pKa = 6.51PLVDD494 pKa = 3.13ISHH497 pKa = 6.45YY498 pKa = 10.31DD499 pKa = 3.36DD500 pKa = 4.04RR501 pKa = 11.84RR502 pKa = 11.84AIEE505 pKa = 4.28NRR507 pKa = 11.84SFMYY511 pKa = 10.08KK512 pKa = 9.92VEE514 pKa = 4.53LGSEE518 pKa = 4.2PVNAHH523 pKa = 5.48IKK525 pKa = 10.03FPNRR529 pKa = 11.84MIPIKK534 pKa = 10.4KK535 pKa = 9.95NPEE538 pKa = 3.49LTQFVLASMQYY549 pKa = 9.65VHH551 pKa = 6.69TNYY554 pKa = 9.66MNRR557 pKa = 11.84PDD559 pKa = 3.86KK560 pKa = 10.74KK561 pKa = 10.69FKK563 pKa = 10.42IGFFNKK569 pKa = 10.31LYY571 pKa = 10.73DD572 pKa = 3.86ALFEE576 pKa = 4.33NSS578 pKa = 3.11
Molecular weight: 68.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1440 |
427 |
578 |
480.0 |
55.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.347 ± 1.66 | 1.528 ± 0.332 |
5.139 ± 0.301 | 7.917 ± 0.914 |
4.167 ± 0.586 | 5.903 ± 0.88 |
2.431 ± 0.53 | 5.694 ± 0.764 |
9.028 ± 0.875 | 6.25 ± 0.425 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.889 ± 0.881 | 7.083 ± 0.265 |
2.986 ± 0.212 | 4.444 ± 0.518 |
5.556 ± 0.694 | 6.111 ± 0.234 |
5.069 ± 0.494 | 5.556 ± 0.391 |
1.736 ± 0.234 | 4.167 ± 0.832 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
