
Hubei sobemo-like virus 36
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.25
Get precalculated fractions of proteins
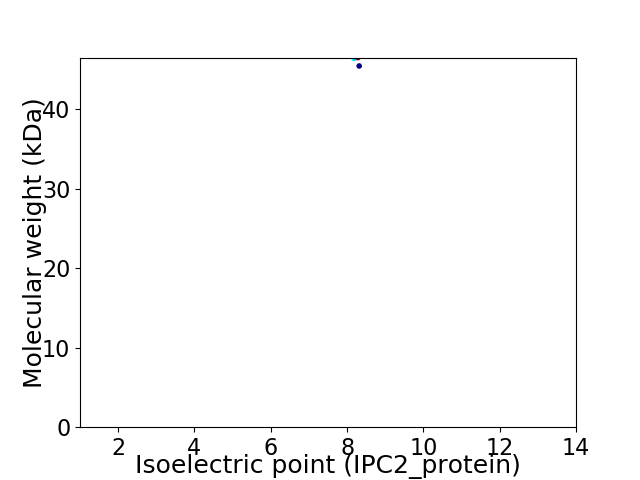
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEM8|A0A1L3KEM8_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 36 OX=1923223 PE=4 SV=1
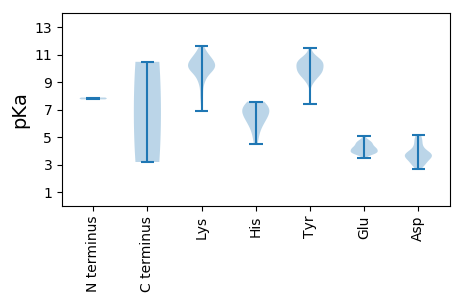
MM1 pKa = 7.87RR2 pKa = 11.84SLLFHH7 pKa = 6.79MKK9 pKa = 9.94RR10 pKa = 11.84HH11 pKa = 6.12IDD13 pKa = 3.47ISDD16 pKa = 3.51LAMDD20 pKa = 4.62LEE22 pKa = 4.52TRR24 pKa = 11.84TRR26 pKa = 11.84VVNKK30 pKa = 9.94ILLDD34 pKa = 3.87LKK36 pKa = 9.26PARR39 pKa = 11.84WEE41 pKa = 3.81IPEE44 pKa = 4.7DD45 pKa = 3.64FLSFSHH51 pKa = 7.55FYY53 pKa = 10.54RR54 pKa = 11.84VVKK57 pKa = 10.69DD58 pKa = 3.48LDD60 pKa = 3.99YY61 pKa = 10.52TSSPGYY67 pKa = 9.84PYY69 pKa = 10.13MRR71 pKa = 11.84TATNIRR77 pKa = 11.84DD78 pKa = 3.79FLKK81 pKa = 10.44VDD83 pKa = 3.69SSGEE87 pKa = 3.8PHH89 pKa = 7.01PEE91 pKa = 3.46KK92 pKa = 10.95LNYY95 pKa = 9.63LWSLVKK101 pKa = 10.75NQIATKK107 pKa = 9.74KK108 pKa = 9.91ADD110 pKa = 4.68PIRR113 pKa = 11.84MFIKK117 pKa = 10.37PEE119 pKa = 4.79PISLTKK125 pKa = 10.84VKK127 pKa = 10.38DD128 pKa = 3.09KK129 pKa = 10.52RR130 pKa = 11.84YY131 pKa = 9.71RR132 pKa = 11.84IISAVSLVDD141 pKa = 3.76QIIDD145 pKa = 3.2HH146 pKa = 6.14MLFANFNHH154 pKa = 6.78KK155 pKa = 10.22LVEE158 pKa = 4.23NCQRR162 pKa = 11.84TPVKK166 pKa = 10.06TGWTPIQGGWKK177 pKa = 9.03VVPRR181 pKa = 11.84LGMVSTDD188 pKa = 2.93KK189 pKa = 11.4SSFDD193 pKa = 3.12WTVRR197 pKa = 11.84PWMLDD202 pKa = 3.07VEE204 pKa = 4.6LEE206 pKa = 3.83IRR208 pKa = 11.84TRR210 pKa = 11.84LCNNVTEE217 pKa = 4.78EE218 pKa = 4.1WKK220 pKa = 10.93QMAKK224 pKa = 8.57WRR226 pKa = 11.84YY227 pKa = 7.42EE228 pKa = 3.74QLFNRR233 pKa = 11.84PLFINSAGIVFRR245 pKa = 11.84SNISGIMKK253 pKa = 9.79SGCVNTISTNSIIQLIIHH271 pKa = 5.79YY272 pKa = 9.73RR273 pKa = 11.84VSAEE277 pKa = 3.82LNQPFPYY284 pKa = 9.2IWAMGDD290 pKa = 3.63DD291 pKa = 4.9VIQEE295 pKa = 4.52DD296 pKa = 3.82SGSDD300 pKa = 3.52YY301 pKa = 9.69FTHH304 pKa = 7.26LEE306 pKa = 4.19RR307 pKa = 11.84YY308 pKa = 9.72CILKK312 pKa = 10.49EE313 pKa = 3.96KK314 pKa = 6.88TTHH317 pKa = 6.14VEE319 pKa = 3.88FAGYY323 pKa = 9.92RR324 pKa = 11.84YY325 pKa = 10.12NGYY328 pKa = 9.4NVEE331 pKa = 3.86PLYY334 pKa = 10.49FGKK337 pKa = 9.87HH338 pKa = 5.46AYY340 pKa = 9.71VLLHH344 pKa = 6.39QKK346 pKa = 9.82LQYY349 pKa = 8.95ITEE352 pKa = 4.23TANAYY357 pKa = 10.13ALLYY361 pKa = 10.1HH362 pKa = 6.65RR363 pKa = 11.84SKK365 pKa = 10.65RR366 pKa = 11.84ASKK369 pKa = 9.91IKK371 pKa = 9.93TYY373 pKa = 10.6LRR375 pKa = 11.84QLNPEE380 pKa = 5.08LISDD384 pKa = 5.03DD385 pKa = 4.15VLDD388 pKa = 4.98DD389 pKa = 3.69LWDD392 pKa = 3.94NEE394 pKa = 4.5SS395 pKa = 3.18
MM1 pKa = 7.87RR2 pKa = 11.84SLLFHH7 pKa = 6.79MKK9 pKa = 9.94RR10 pKa = 11.84HH11 pKa = 6.12IDD13 pKa = 3.47ISDD16 pKa = 3.51LAMDD20 pKa = 4.62LEE22 pKa = 4.52TRR24 pKa = 11.84TRR26 pKa = 11.84VVNKK30 pKa = 9.94ILLDD34 pKa = 3.87LKK36 pKa = 9.26PARR39 pKa = 11.84WEE41 pKa = 3.81IPEE44 pKa = 4.7DD45 pKa = 3.64FLSFSHH51 pKa = 7.55FYY53 pKa = 10.54RR54 pKa = 11.84VVKK57 pKa = 10.69DD58 pKa = 3.48LDD60 pKa = 3.99YY61 pKa = 10.52TSSPGYY67 pKa = 9.84PYY69 pKa = 10.13MRR71 pKa = 11.84TATNIRR77 pKa = 11.84DD78 pKa = 3.79FLKK81 pKa = 10.44VDD83 pKa = 3.69SSGEE87 pKa = 3.8PHH89 pKa = 7.01PEE91 pKa = 3.46KK92 pKa = 10.95LNYY95 pKa = 9.63LWSLVKK101 pKa = 10.75NQIATKK107 pKa = 9.74KK108 pKa = 9.91ADD110 pKa = 4.68PIRR113 pKa = 11.84MFIKK117 pKa = 10.37PEE119 pKa = 4.79PISLTKK125 pKa = 10.84VKK127 pKa = 10.38DD128 pKa = 3.09KK129 pKa = 10.52RR130 pKa = 11.84YY131 pKa = 9.71RR132 pKa = 11.84IISAVSLVDD141 pKa = 3.76QIIDD145 pKa = 3.2HH146 pKa = 6.14MLFANFNHH154 pKa = 6.78KK155 pKa = 10.22LVEE158 pKa = 4.23NCQRR162 pKa = 11.84TPVKK166 pKa = 10.06TGWTPIQGGWKK177 pKa = 9.03VVPRR181 pKa = 11.84LGMVSTDD188 pKa = 2.93KK189 pKa = 11.4SSFDD193 pKa = 3.12WTVRR197 pKa = 11.84PWMLDD202 pKa = 3.07VEE204 pKa = 4.6LEE206 pKa = 3.83IRR208 pKa = 11.84TRR210 pKa = 11.84LCNNVTEE217 pKa = 4.78EE218 pKa = 4.1WKK220 pKa = 10.93QMAKK224 pKa = 8.57WRR226 pKa = 11.84YY227 pKa = 7.42EE228 pKa = 3.74QLFNRR233 pKa = 11.84PLFINSAGIVFRR245 pKa = 11.84SNISGIMKK253 pKa = 9.79SGCVNTISTNSIIQLIIHH271 pKa = 5.79YY272 pKa = 9.73RR273 pKa = 11.84VSAEE277 pKa = 3.82LNQPFPYY284 pKa = 9.2IWAMGDD290 pKa = 3.63DD291 pKa = 4.9VIQEE295 pKa = 4.52DD296 pKa = 3.82SGSDD300 pKa = 3.52YY301 pKa = 9.69FTHH304 pKa = 7.26LEE306 pKa = 4.19RR307 pKa = 11.84YY308 pKa = 9.72CILKK312 pKa = 10.49EE313 pKa = 3.96KK314 pKa = 6.88TTHH317 pKa = 6.14VEE319 pKa = 3.88FAGYY323 pKa = 9.92RR324 pKa = 11.84YY325 pKa = 10.12NGYY328 pKa = 9.4NVEE331 pKa = 3.86PLYY334 pKa = 10.49FGKK337 pKa = 9.87HH338 pKa = 5.46AYY340 pKa = 9.71VLLHH344 pKa = 6.39QKK346 pKa = 9.82LQYY349 pKa = 8.95ITEE352 pKa = 4.23TANAYY357 pKa = 10.13ALLYY361 pKa = 10.1HH362 pKa = 6.65RR363 pKa = 11.84SKK365 pKa = 10.65RR366 pKa = 11.84ASKK369 pKa = 9.91IKK371 pKa = 9.93TYY373 pKa = 10.6LRR375 pKa = 11.84QLNPEE380 pKa = 5.08LISDD384 pKa = 5.03DD385 pKa = 4.15VLDD388 pKa = 4.98DD389 pKa = 3.69LWDD392 pKa = 3.94NEE394 pKa = 4.5SS395 pKa = 3.18
Molecular weight: 46.36 kDa
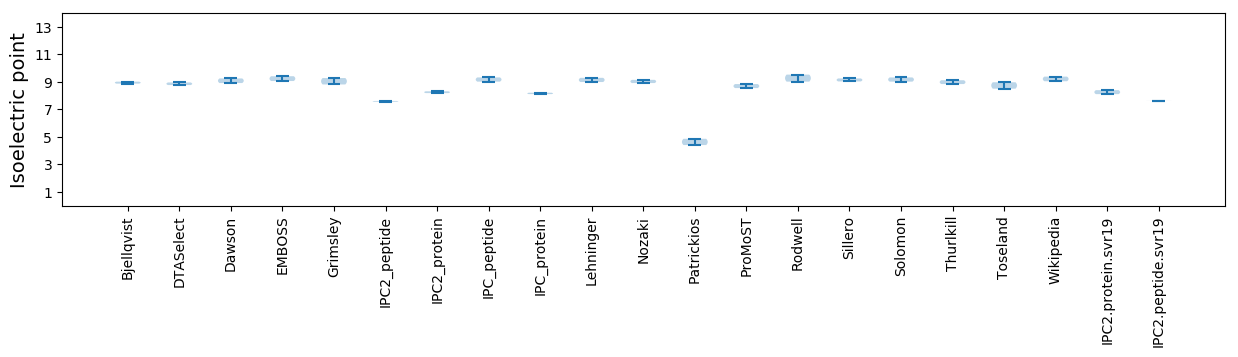
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEM8|A0A1L3KEM8_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 36 OX=1923223 PE=4 SV=1
MM1 pKa = 7.74PLHH4 pKa = 7.38VYY6 pKa = 9.72RR7 pKa = 11.84QAVPDD12 pKa = 3.57ILIGKK17 pKa = 8.11NGSYY21 pKa = 10.72FEE23 pKa = 4.42ISNAPMIEE31 pKa = 4.07SDD33 pKa = 4.46SVTDD37 pKa = 3.7LCYY40 pKa = 10.44VTVKK44 pKa = 10.3PQIFSQLKK52 pKa = 7.73TSIARR57 pKa = 11.84FPKK60 pKa = 10.19GMVEE64 pKa = 4.2GFVSAVGRR72 pKa = 11.84SGASQGSLEE81 pKa = 4.2KK82 pKa = 10.37AASRR86 pKa = 11.84GIVIYY91 pKa = 10.78NGSTKK96 pKa = 10.12PGYY99 pKa = 10.3SGAAYY104 pKa = 10.52LIGNQIYY111 pKa = 10.38GMHH114 pKa = 6.92IGHH117 pKa = 6.65SSGANVGISHH127 pKa = 7.06VVLQYY132 pKa = 11.49DD133 pKa = 3.73LNKK136 pKa = 10.1YY137 pKa = 9.1KK138 pKa = 11.1YY139 pKa = 9.44HH140 pKa = 5.12VTVDD144 pKa = 3.43KK145 pKa = 10.66EE146 pKa = 4.4GSVTFSSPGMMGSKK160 pKa = 10.48EE161 pKa = 3.82EE162 pKa = 4.25DD163 pKa = 3.01LTRR166 pKa = 11.84VYY168 pKa = 10.68DD169 pKa = 4.34GKK171 pKa = 10.16WSADD175 pKa = 2.93EE176 pKa = 4.04FLRR179 pKa = 11.84EE180 pKa = 3.92RR181 pKa = 11.84KK182 pKa = 9.3GLKK185 pKa = 8.95SWNDD189 pKa = 2.71IKK191 pKa = 11.33LDD193 pKa = 5.12DD194 pKa = 4.03KK195 pKa = 11.62ALWANEE201 pKa = 3.89FEE203 pKa = 4.56STLRR207 pKa = 11.84SLPPDD212 pKa = 3.25VLKK215 pKa = 10.83FVVDD219 pKa = 3.68RR220 pKa = 11.84AGAEE224 pKa = 4.1LQKK227 pKa = 11.43SNITVQGQSDD237 pKa = 4.06DD238 pKa = 4.55EE239 pKa = 4.68IPVDD243 pKa = 2.92IHH245 pKa = 7.58IYY247 pKa = 10.37SPQQQEE253 pKa = 4.32SLSQRR258 pKa = 11.84VTNLEE263 pKa = 3.71ARR265 pKa = 11.84VSKK268 pKa = 10.97LEE270 pKa = 3.88QKK272 pKa = 10.13KK273 pKa = 10.26APKK276 pKa = 9.53QGPVQKK282 pKa = 9.69VTVFKK287 pKa = 10.9CLQCDD292 pKa = 3.68STSTTRR298 pKa = 11.84LAVANHH304 pKa = 7.25MITKK308 pKa = 9.18HH309 pKa = 4.5QVPEE313 pKa = 3.91EE314 pKa = 4.06SLDD317 pKa = 3.67LGAILAMEE325 pKa = 4.66KK326 pKa = 9.91EE327 pKa = 4.48VKK329 pKa = 10.25PEE331 pKa = 3.81GVLSSDD337 pKa = 3.56EE338 pKa = 4.18SDD340 pKa = 3.33LVGNGKK346 pKa = 8.0VFRR349 pKa = 11.84NRR351 pKa = 11.84SLSPPKK357 pKa = 10.08KK358 pKa = 9.8SSNSKK363 pKa = 10.21SSSSPSRR370 pKa = 11.84KK371 pKa = 9.14PNRR374 pKa = 11.84FQQSAQNQKK383 pKa = 10.6NMEE386 pKa = 4.17NLMNKK391 pKa = 9.95LNEE394 pKa = 4.11NLLKK398 pKa = 10.63LVQNLPGPSSAQGQKK413 pKa = 10.47
MM1 pKa = 7.74PLHH4 pKa = 7.38VYY6 pKa = 9.72RR7 pKa = 11.84QAVPDD12 pKa = 3.57ILIGKK17 pKa = 8.11NGSYY21 pKa = 10.72FEE23 pKa = 4.42ISNAPMIEE31 pKa = 4.07SDD33 pKa = 4.46SVTDD37 pKa = 3.7LCYY40 pKa = 10.44VTVKK44 pKa = 10.3PQIFSQLKK52 pKa = 7.73TSIARR57 pKa = 11.84FPKK60 pKa = 10.19GMVEE64 pKa = 4.2GFVSAVGRR72 pKa = 11.84SGASQGSLEE81 pKa = 4.2KK82 pKa = 10.37AASRR86 pKa = 11.84GIVIYY91 pKa = 10.78NGSTKK96 pKa = 10.12PGYY99 pKa = 10.3SGAAYY104 pKa = 10.52LIGNQIYY111 pKa = 10.38GMHH114 pKa = 6.92IGHH117 pKa = 6.65SSGANVGISHH127 pKa = 7.06VVLQYY132 pKa = 11.49DD133 pKa = 3.73LNKK136 pKa = 10.1YY137 pKa = 9.1KK138 pKa = 11.1YY139 pKa = 9.44HH140 pKa = 5.12VTVDD144 pKa = 3.43KK145 pKa = 10.66EE146 pKa = 4.4GSVTFSSPGMMGSKK160 pKa = 10.48EE161 pKa = 3.82EE162 pKa = 4.25DD163 pKa = 3.01LTRR166 pKa = 11.84VYY168 pKa = 10.68DD169 pKa = 4.34GKK171 pKa = 10.16WSADD175 pKa = 2.93EE176 pKa = 4.04FLRR179 pKa = 11.84EE180 pKa = 3.92RR181 pKa = 11.84KK182 pKa = 9.3GLKK185 pKa = 8.95SWNDD189 pKa = 2.71IKK191 pKa = 11.33LDD193 pKa = 5.12DD194 pKa = 4.03KK195 pKa = 11.62ALWANEE201 pKa = 3.89FEE203 pKa = 4.56STLRR207 pKa = 11.84SLPPDD212 pKa = 3.25VLKK215 pKa = 10.83FVVDD219 pKa = 3.68RR220 pKa = 11.84AGAEE224 pKa = 4.1LQKK227 pKa = 11.43SNITVQGQSDD237 pKa = 4.06DD238 pKa = 4.55EE239 pKa = 4.68IPVDD243 pKa = 2.92IHH245 pKa = 7.58IYY247 pKa = 10.37SPQQQEE253 pKa = 4.32SLSQRR258 pKa = 11.84VTNLEE263 pKa = 3.71ARR265 pKa = 11.84VSKK268 pKa = 10.97LEE270 pKa = 3.88QKK272 pKa = 10.13KK273 pKa = 10.26APKK276 pKa = 9.53QGPVQKK282 pKa = 9.69VTVFKK287 pKa = 10.9CLQCDD292 pKa = 3.68STSTTRR298 pKa = 11.84LAVANHH304 pKa = 7.25MITKK308 pKa = 9.18HH309 pKa = 4.5QVPEE313 pKa = 3.91EE314 pKa = 4.06SLDD317 pKa = 3.67LGAILAMEE325 pKa = 4.66KK326 pKa = 9.91EE327 pKa = 4.48VKK329 pKa = 10.25PEE331 pKa = 3.81GVLSSDD337 pKa = 3.56EE338 pKa = 4.18SDD340 pKa = 3.33LVGNGKK346 pKa = 8.0VFRR349 pKa = 11.84NRR351 pKa = 11.84SLSPPKK357 pKa = 10.08KK358 pKa = 9.8SSNSKK363 pKa = 10.21SSSSPSRR370 pKa = 11.84KK371 pKa = 9.14PNRR374 pKa = 11.84FQQSAQNQKK383 pKa = 10.6NMEE386 pKa = 4.17NLMNKK391 pKa = 9.95LNEE394 pKa = 4.11NLLKK398 pKa = 10.63LVQNLPGPSSAQGQKK413 pKa = 10.47
Molecular weight: 45.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
808 |
395 |
413 |
404.0 |
45.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.95 ± 0.437 | 0.866 ± 0.099 |
5.569 ± 0.514 | 5.569 ± 0.171 |
3.342 ± 0.479 | 5.446 ± 1.285 |
2.475 ± 0.38 | 6.064 ± 1.035 |
7.673 ± 0.566 | 8.911 ± 0.48 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.126 | 5.198 ± 0.091 |
4.827 ± 0.182 | 4.579 ± 1.042 |
5.074 ± 0.848 | 9.53 ± 1.65 |
4.579 ± 0.67 | 7.178 ± 0.574 |
1.609 ± 0.624 | 3.96 ± 0.746 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
