
Galliform aveparvovirus 1 (isolate Chicken/Hungary/ABU-P1/-/-) (ChPV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Aveparvovirus; Galliform aveparvovirus 1
Average proteome isoelectric point is 5.17
Get precalculated fractions of proteins
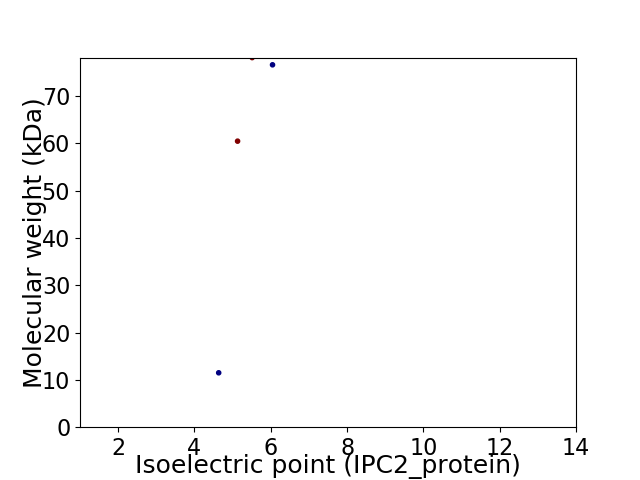
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
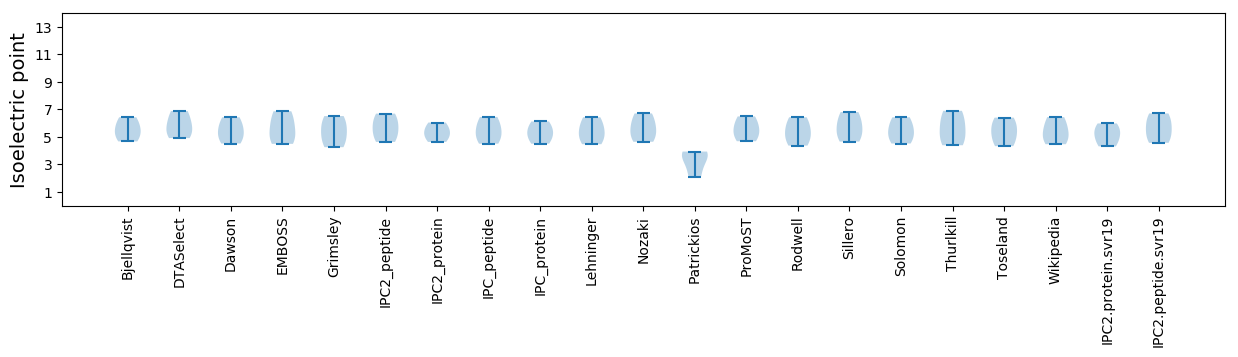
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D3X6W9|D3X6W9_CHPV1 VP1 OS=Galliform aveparvovirus 1 (isolate Chicken/Hungary/ABU-P1/-/-) OX=717641 GN=VP1 PE=3 SV=1
MM1 pKa = 7.54CWYY4 pKa = 9.58NANITQIQTVEE15 pKa = 4.34GDD17 pKa = 3.29PCTNPDD23 pKa = 3.7VFHH26 pKa = 7.01EE27 pKa = 4.94LYY29 pKa = 10.8FPIKK33 pKa = 9.05QHH35 pKa = 6.94AMDD38 pKa = 4.3ARR40 pKa = 11.84MLKK43 pKa = 9.56MKK45 pKa = 10.35IEE47 pKa = 4.43DD48 pKa = 4.56KK49 pKa = 10.43IQKK52 pKa = 10.38GDD54 pKa = 3.54FTQDD58 pKa = 2.11IHH60 pKa = 9.4LDD62 pKa = 3.55MTLTQCIQTALSDD75 pKa = 4.11DD76 pKa = 4.07PAGIWPCVDD85 pKa = 3.88RR86 pKa = 11.84LANYY90 pKa = 10.1CNVILASIGTQQ101 pKa = 2.95
MM1 pKa = 7.54CWYY4 pKa = 9.58NANITQIQTVEE15 pKa = 4.34GDD17 pKa = 3.29PCTNPDD23 pKa = 3.7VFHH26 pKa = 7.01EE27 pKa = 4.94LYY29 pKa = 10.8FPIKK33 pKa = 9.05QHH35 pKa = 6.94AMDD38 pKa = 4.3ARR40 pKa = 11.84MLKK43 pKa = 9.56MKK45 pKa = 10.35IEE47 pKa = 4.43DD48 pKa = 4.56KK49 pKa = 10.43IQKK52 pKa = 10.38GDD54 pKa = 3.54FTQDD58 pKa = 2.11IHH60 pKa = 9.4LDD62 pKa = 3.55MTLTQCIQTALSDD75 pKa = 4.11DD76 pKa = 4.07PAGIWPCVDD85 pKa = 3.88RR86 pKa = 11.84LANYY90 pKa = 10.1CNVILASIGTQQ101 pKa = 2.95
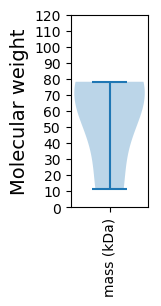
Molecular weight: 11.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D3X6X0|D3X6X0_CHPV1 VP2 OS=Galliform aveparvovirus 1 (isolate Chicken/Hungary/ABU-P1/-/-) OX=717641 GN=VP2 PE=3 SV=1
MM1 pKa = 7.73KK2 pKa = 10.34YY3 pKa = 9.62KK4 pKa = 10.54APKK7 pKa = 9.99GYY9 pKa = 10.38VPSLPTTDD17 pKa = 3.26EE18 pKa = 3.84EE19 pKa = 5.11AYY21 pKa = 10.57HH22 pKa = 5.48HH23 pKa = 5.6TRR25 pKa = 11.84WWLDD29 pKa = 3.09HH30 pKa = 6.06VEE32 pKa = 4.21NYY34 pKa = 9.84GKK36 pKa = 10.32NKK38 pKa = 10.47AIGRR42 pKa = 11.84GKK44 pKa = 10.37GKK46 pKa = 10.08ARR48 pKa = 11.84KK49 pKa = 8.92EE50 pKa = 3.58LTPQQKK56 pKa = 10.52AEE58 pKa = 4.06RR59 pKa = 11.84KK60 pKa = 9.73RR61 pKa = 11.84FFITQAQKK69 pKa = 11.0NKK71 pKa = 9.36KK72 pKa = 8.18PNYY75 pKa = 8.63GIPSSSQHH83 pKa = 6.38KK84 pKa = 10.15EE85 pKa = 3.51FFKK88 pKa = 10.88NHH90 pKa = 5.37QGAQGGKK97 pKa = 9.06AKK99 pKa = 10.48QKK101 pKa = 9.22TKK103 pKa = 10.32HH104 pKa = 6.26IEE106 pKa = 3.78NQAPQRR112 pKa = 11.84QEE114 pKa = 4.02DD115 pKa = 4.5PEE117 pKa = 4.44EE118 pKa = 4.84GPAPKK123 pKa = 9.94QPRR126 pKa = 11.84VEE128 pKa = 4.13EE129 pKa = 3.82NPYY132 pKa = 9.72NQEE135 pKa = 3.76EE136 pKa = 4.7LEE138 pKa = 4.14EE139 pKa = 4.18MMADD143 pKa = 3.59EE144 pKa = 5.22NEE146 pKa = 4.24PDD148 pKa = 3.26QSGIPMEE155 pKa = 5.07LSDD158 pKa = 5.16GPGTVGGGGGGGIGHH173 pKa = 6.08STGNWNCDD181 pKa = 3.88TIWAEE186 pKa = 4.35NTCKK190 pKa = 10.12TYY192 pKa = 11.09ASRR195 pKa = 11.84HH196 pKa = 5.21CVCLMRR202 pKa = 11.84DD203 pKa = 3.08MDD205 pKa = 3.74QYY207 pKa = 11.7KK208 pKa = 10.82AIGNANGQHH217 pKa = 6.56GLDD220 pKa = 4.19TEE222 pKa = 4.3NQTQYY227 pKa = 11.33FGFSTPWNYY236 pKa = 9.77IDD238 pKa = 4.04FNQYY242 pKa = 9.58SVHH245 pKa = 6.49FSPRR249 pKa = 11.84DD250 pKa = 3.35WQQLVNNYY258 pKa = 9.49SRR260 pKa = 11.84WRR262 pKa = 11.84PRR264 pKa = 11.84AVHH267 pKa = 6.1VKK269 pKa = 9.8IFNLQVIQKK278 pKa = 7.67TVTDD282 pKa = 3.47SGTQYY287 pKa = 11.55SNDD290 pKa = 3.3LTGTIQIFADD300 pKa = 3.29QEE302 pKa = 3.86GRR304 pKa = 11.84YY305 pKa = 8.96PRR307 pKa = 11.84LLYY310 pKa = 10.11PNQTTLMGPFPSNIYY325 pKa = 9.89YY326 pKa = 10.39LPQYY330 pKa = 11.35AYY332 pKa = 6.86MTSMFNSGGPTVSSWWGFPTRR353 pKa = 11.84EE354 pKa = 3.94SAFYY358 pKa = 10.8CLDD361 pKa = 3.62EE362 pKa = 5.5SPSQMLRR369 pKa = 11.84TGNEE373 pKa = 3.5WEE375 pKa = 4.34TTFIFPSSTQWCSNKK390 pKa = 9.53ISTVSITQRR399 pKa = 11.84QNPLYY404 pKa = 9.56DD405 pKa = 3.09TWNVNGRR412 pKa = 11.84GDD414 pKa = 3.5DD415 pKa = 3.43AKK417 pKa = 10.82RR418 pKa = 11.84GNFCTWRR425 pKa = 11.84SPWYY429 pKa = 8.08PGPYY433 pKa = 9.04IYY435 pKa = 10.27LTDD438 pKa = 3.69TSAASQRR445 pKa = 11.84LEE447 pKa = 3.71NLANVAVGPDD457 pKa = 4.01GMPLAPGMPQYY468 pKa = 10.59RR469 pKa = 11.84SEE471 pKa = 4.16SDD473 pKa = 2.88KK474 pKa = 11.71DD475 pKa = 3.75EE476 pKa = 4.33YY477 pKa = 11.7LHH479 pKa = 6.2TFWVPKK485 pKa = 8.65KK486 pKa = 10.66VKK488 pKa = 9.85MNEE491 pKa = 3.38GDD493 pKa = 3.24IRR495 pKa = 11.84NAQIDD500 pKa = 3.9VSTATKK506 pKa = 10.3VQVDD510 pKa = 3.66TSTLYY515 pKa = 8.65NTCPRR520 pKa = 11.84SGYY523 pKa = 9.47MSTVSQGGAPAPIVWSGCVPGMIWDD548 pKa = 4.13NRR550 pKa = 11.84PATYY554 pKa = 10.12FDD556 pKa = 5.85PIWQQFPEE564 pKa = 4.05TDD566 pKa = 3.39EE567 pKa = 3.85QFKK570 pKa = 10.7IISQLGGIPVKK581 pKa = 10.2EE582 pKa = 4.33SPGHH586 pKa = 5.77IFVKK590 pKa = 8.93VTPKK594 pKa = 9.21PTGAANGLIDD604 pKa = 4.38EE605 pKa = 4.89YY606 pKa = 11.14CTFTCTVEE614 pKa = 4.23IEE616 pKa = 4.34WEE618 pKa = 4.11LEE620 pKa = 3.66PLNTHH625 pKa = 5.84RR626 pKa = 11.84WNMRR630 pKa = 11.84SMISYY635 pKa = 7.73EE636 pKa = 4.11TTDD639 pKa = 3.03AKK641 pKa = 10.71AGRR644 pKa = 11.84QIVDD648 pKa = 3.27EE649 pKa = 4.36NGQYY653 pKa = 9.53QVNVNSADD661 pKa = 3.08ITRR664 pKa = 11.84LYY666 pKa = 6.64MTKK669 pKa = 10.31RR670 pKa = 11.84APRR673 pKa = 11.84TNN675 pKa = 3.16
MM1 pKa = 7.73KK2 pKa = 10.34YY3 pKa = 9.62KK4 pKa = 10.54APKK7 pKa = 9.99GYY9 pKa = 10.38VPSLPTTDD17 pKa = 3.26EE18 pKa = 3.84EE19 pKa = 5.11AYY21 pKa = 10.57HH22 pKa = 5.48HH23 pKa = 5.6TRR25 pKa = 11.84WWLDD29 pKa = 3.09HH30 pKa = 6.06VEE32 pKa = 4.21NYY34 pKa = 9.84GKK36 pKa = 10.32NKK38 pKa = 10.47AIGRR42 pKa = 11.84GKK44 pKa = 10.37GKK46 pKa = 10.08ARR48 pKa = 11.84KK49 pKa = 8.92EE50 pKa = 3.58LTPQQKK56 pKa = 10.52AEE58 pKa = 4.06RR59 pKa = 11.84KK60 pKa = 9.73RR61 pKa = 11.84FFITQAQKK69 pKa = 11.0NKK71 pKa = 9.36KK72 pKa = 8.18PNYY75 pKa = 8.63GIPSSSQHH83 pKa = 6.38KK84 pKa = 10.15EE85 pKa = 3.51FFKK88 pKa = 10.88NHH90 pKa = 5.37QGAQGGKK97 pKa = 9.06AKK99 pKa = 10.48QKK101 pKa = 9.22TKK103 pKa = 10.32HH104 pKa = 6.26IEE106 pKa = 3.78NQAPQRR112 pKa = 11.84QEE114 pKa = 4.02DD115 pKa = 4.5PEE117 pKa = 4.44EE118 pKa = 4.84GPAPKK123 pKa = 9.94QPRR126 pKa = 11.84VEE128 pKa = 4.13EE129 pKa = 3.82NPYY132 pKa = 9.72NQEE135 pKa = 3.76EE136 pKa = 4.7LEE138 pKa = 4.14EE139 pKa = 4.18MMADD143 pKa = 3.59EE144 pKa = 5.22NEE146 pKa = 4.24PDD148 pKa = 3.26QSGIPMEE155 pKa = 5.07LSDD158 pKa = 5.16GPGTVGGGGGGGIGHH173 pKa = 6.08STGNWNCDD181 pKa = 3.88TIWAEE186 pKa = 4.35NTCKK190 pKa = 10.12TYY192 pKa = 11.09ASRR195 pKa = 11.84HH196 pKa = 5.21CVCLMRR202 pKa = 11.84DD203 pKa = 3.08MDD205 pKa = 3.74QYY207 pKa = 11.7KK208 pKa = 10.82AIGNANGQHH217 pKa = 6.56GLDD220 pKa = 4.19TEE222 pKa = 4.3NQTQYY227 pKa = 11.33FGFSTPWNYY236 pKa = 9.77IDD238 pKa = 4.04FNQYY242 pKa = 9.58SVHH245 pKa = 6.49FSPRR249 pKa = 11.84DD250 pKa = 3.35WQQLVNNYY258 pKa = 9.49SRR260 pKa = 11.84WRR262 pKa = 11.84PRR264 pKa = 11.84AVHH267 pKa = 6.1VKK269 pKa = 9.8IFNLQVIQKK278 pKa = 7.67TVTDD282 pKa = 3.47SGTQYY287 pKa = 11.55SNDD290 pKa = 3.3LTGTIQIFADD300 pKa = 3.29QEE302 pKa = 3.86GRR304 pKa = 11.84YY305 pKa = 8.96PRR307 pKa = 11.84LLYY310 pKa = 10.11PNQTTLMGPFPSNIYY325 pKa = 9.89YY326 pKa = 10.39LPQYY330 pKa = 11.35AYY332 pKa = 6.86MTSMFNSGGPTVSSWWGFPTRR353 pKa = 11.84EE354 pKa = 3.94SAFYY358 pKa = 10.8CLDD361 pKa = 3.62EE362 pKa = 5.5SPSQMLRR369 pKa = 11.84TGNEE373 pKa = 3.5WEE375 pKa = 4.34TTFIFPSSTQWCSNKK390 pKa = 9.53ISTVSITQRR399 pKa = 11.84QNPLYY404 pKa = 9.56DD405 pKa = 3.09TWNVNGRR412 pKa = 11.84GDD414 pKa = 3.5DD415 pKa = 3.43AKK417 pKa = 10.82RR418 pKa = 11.84GNFCTWRR425 pKa = 11.84SPWYY429 pKa = 8.08PGPYY433 pKa = 9.04IYY435 pKa = 10.27LTDD438 pKa = 3.69TSAASQRR445 pKa = 11.84LEE447 pKa = 3.71NLANVAVGPDD457 pKa = 4.01GMPLAPGMPQYY468 pKa = 10.59RR469 pKa = 11.84SEE471 pKa = 4.16SDD473 pKa = 2.88KK474 pKa = 11.71DD475 pKa = 3.75EE476 pKa = 4.33YY477 pKa = 11.7LHH479 pKa = 6.2TFWVPKK485 pKa = 8.65KK486 pKa = 10.66VKK488 pKa = 9.85MNEE491 pKa = 3.38GDD493 pKa = 3.24IRR495 pKa = 11.84NAQIDD500 pKa = 3.9VSTATKK506 pKa = 10.3VQVDD510 pKa = 3.66TSTLYY515 pKa = 8.65NTCPRR520 pKa = 11.84SGYY523 pKa = 9.47MSTVSQGGAPAPIVWSGCVPGMIWDD548 pKa = 4.13NRR550 pKa = 11.84PATYY554 pKa = 10.12FDD556 pKa = 5.85PIWQQFPEE564 pKa = 4.05TDD566 pKa = 3.39EE567 pKa = 3.85QFKK570 pKa = 10.7IISQLGGIPVKK581 pKa = 10.2EE582 pKa = 4.33SPGHH586 pKa = 5.77IFVKK590 pKa = 8.93VTPKK594 pKa = 9.21PTGAANGLIDD604 pKa = 4.38EE605 pKa = 4.89YY606 pKa = 11.14CTFTCTVEE614 pKa = 4.23IEE616 pKa = 4.34WEE618 pKa = 4.11LEE620 pKa = 3.66PLNTHH625 pKa = 5.84RR626 pKa = 11.84WNMRR630 pKa = 11.84SMISYY635 pKa = 7.73EE636 pKa = 4.11TTDD639 pKa = 3.03AKK641 pKa = 10.71AGRR644 pKa = 11.84QIVDD648 pKa = 3.27EE649 pKa = 4.36NGQYY653 pKa = 9.53QVNVNSADD661 pKa = 3.08ITRR664 pKa = 11.84LYY666 pKa = 6.64MTKK669 pKa = 10.31RR670 pKa = 11.84APRR673 pKa = 11.84TNN675 pKa = 3.16
Molecular weight: 76.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2006 |
101 |
694 |
501.5 |
56.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.334 ± 0.19 | 2.542 ± 0.434 |
5.234 ± 0.415 | 5.833 ± 0.663 |
3.838 ± 0.292 | 7.228 ± 0.521 |
2.044 ± 0.206 | 5.583 ± 0.407 |
5.135 ± 0.771 | 5.982 ± 1.107 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.212 | 6.231 ± 0.138 |
6.78 ± 0.206 | 5.384 ± 0.683 |
4.187 ± 0.307 | 6.73 ± 0.429 |
7.428 ± 0.76 | 4.786 ± 0.171 |
2.742 ± 0.273 | 4.088 ± 0.556 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
