
Cellulomonas sp. HLT2-17
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; unclassified Cellulomonas
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
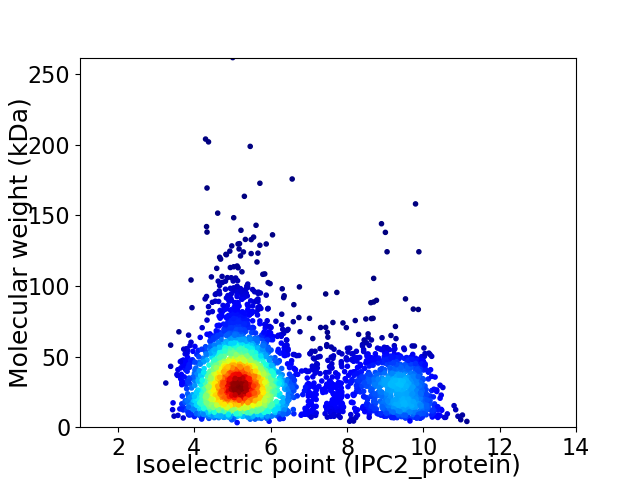
Virtual 2D-PAGE plot for 3785 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q5N5T6|A0A4Q5N5T6_9CELL Efflux RND transporter periplasmic adaptor subunit OS=Cellulomonas sp. HLT2-17 OX=1259133 GN=EUA98_07850 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.33RR3 pKa = 11.84PLVVLTATLAAAGMLAACSGGTTTPKK29 pKa = 10.73GADD32 pKa = 3.22GTAASGPIKK41 pKa = 9.89IWYY44 pKa = 8.93SNNPNEE50 pKa = 4.46VAWGEE55 pKa = 4.17QVVEE59 pKa = 4.18AWNADD64 pKa = 3.54HH65 pKa = 7.65ADD67 pKa = 3.77EE68 pKa = 4.37QVTGQEE74 pKa = 3.89IPAGRR79 pKa = 11.84SSEE82 pKa = 4.19EE83 pKa = 3.91VIGAAITAGNTPCLIYY99 pKa = 9.95NTAPVAVPGFQKK111 pKa = 10.58QGGLVDD117 pKa = 5.48LSTFPDD123 pKa = 3.64GAEE126 pKa = 4.01YY127 pKa = 10.35IEE129 pKa = 4.54ARR131 pKa = 11.84TGEE134 pKa = 4.13AASQYY139 pKa = 10.68VSADD143 pKa = 2.6GDD145 pKa = 4.33YY146 pKa = 11.5YY147 pKa = 11.01QMPWKK152 pKa = 10.03SNPVMFFYY160 pKa = 11.15NKK162 pKa = 10.62AIMTAAGVDD171 pKa = 3.9PEE173 pKa = 4.32NPPLATYY180 pKa = 10.84DD181 pKa = 3.64EE182 pKa = 4.76FLDD185 pKa = 3.71TSRR188 pKa = 11.84KK189 pKa = 9.06IVASGAATYY198 pKa = 10.24AIYY201 pKa = 9.43PSPASEE207 pKa = 5.94FYY209 pKa = 10.87QSWFDD214 pKa = 4.35FYY216 pKa = 10.86PLYY219 pKa = 10.67AAEE222 pKa = 4.56SGGTLLVEE230 pKa = 5.3DD231 pKa = 4.85GASTFAGEE239 pKa = 4.31EE240 pKa = 4.28GTAVADD246 pKa = 4.99FWRR249 pKa = 11.84TIYY252 pKa = 10.48DD253 pKa = 3.33EE254 pKa = 4.51KK255 pKa = 11.22LAGNEE260 pKa = 4.49PYY262 pKa = 10.43TGDD265 pKa = 3.53AFNDD269 pKa = 3.94GVAAIASAGPWAVEE283 pKa = 4.24SYY285 pKa = 11.63GDD287 pKa = 4.05IDD289 pKa = 3.69WGVAPIPTSTGTAPEE304 pKa = 4.02EE305 pKa = 4.53TYY307 pKa = 10.84TFADD311 pKa = 3.72SKK313 pKa = 11.02NVGMYY318 pKa = 10.08SSCEE322 pKa = 3.84NTGTAWEE329 pKa = 4.1FLKK332 pKa = 11.05FSTSEE337 pKa = 4.09EE338 pKa = 3.9QDD340 pKa = 3.14GTLLEE345 pKa = 4.66TTGQMPLRR353 pKa = 11.84ANLTDD358 pKa = 4.15AFPDD362 pKa = 3.78YY363 pKa = 10.64FATNPDD369 pKa = 3.45YY370 pKa = 8.44TTFADD375 pKa = 3.6QANRR379 pKa = 11.84TVDD382 pKa = 3.4VPNVEE387 pKa = 4.13NSVEE391 pKa = 3.97VWQTFRR397 pKa = 11.84DD398 pKa = 3.51AWTSAVIFGEE408 pKa = 4.07DD409 pKa = 4.23EE410 pKa = 3.93IPSALKK416 pKa = 10.56DD417 pKa = 3.34AAGTIDD423 pKa = 3.58EE424 pKa = 4.82LAVKK428 pKa = 10.14KK429 pKa = 10.7
MM1 pKa = 7.38KK2 pKa = 10.33RR3 pKa = 11.84PLVVLTATLAAAGMLAACSGGTTTPKK29 pKa = 10.73GADD32 pKa = 3.22GTAASGPIKK41 pKa = 9.89IWYY44 pKa = 8.93SNNPNEE50 pKa = 4.46VAWGEE55 pKa = 4.17QVVEE59 pKa = 4.18AWNADD64 pKa = 3.54HH65 pKa = 7.65ADD67 pKa = 3.77EE68 pKa = 4.37QVTGQEE74 pKa = 3.89IPAGRR79 pKa = 11.84SSEE82 pKa = 4.19EE83 pKa = 3.91VIGAAITAGNTPCLIYY99 pKa = 9.95NTAPVAVPGFQKK111 pKa = 10.58QGGLVDD117 pKa = 5.48LSTFPDD123 pKa = 3.64GAEE126 pKa = 4.01YY127 pKa = 10.35IEE129 pKa = 4.54ARR131 pKa = 11.84TGEE134 pKa = 4.13AASQYY139 pKa = 10.68VSADD143 pKa = 2.6GDD145 pKa = 4.33YY146 pKa = 11.5YY147 pKa = 11.01QMPWKK152 pKa = 10.03SNPVMFFYY160 pKa = 11.15NKK162 pKa = 10.62AIMTAAGVDD171 pKa = 3.9PEE173 pKa = 4.32NPPLATYY180 pKa = 10.84DD181 pKa = 3.64EE182 pKa = 4.76FLDD185 pKa = 3.71TSRR188 pKa = 11.84KK189 pKa = 9.06IVASGAATYY198 pKa = 10.24AIYY201 pKa = 9.43PSPASEE207 pKa = 5.94FYY209 pKa = 10.87QSWFDD214 pKa = 4.35FYY216 pKa = 10.86PLYY219 pKa = 10.67AAEE222 pKa = 4.56SGGTLLVEE230 pKa = 5.3DD231 pKa = 4.85GASTFAGEE239 pKa = 4.31EE240 pKa = 4.28GTAVADD246 pKa = 4.99FWRR249 pKa = 11.84TIYY252 pKa = 10.48DD253 pKa = 3.33EE254 pKa = 4.51KK255 pKa = 11.22LAGNEE260 pKa = 4.49PYY262 pKa = 10.43TGDD265 pKa = 3.53AFNDD269 pKa = 3.94GVAAIASAGPWAVEE283 pKa = 4.24SYY285 pKa = 11.63GDD287 pKa = 4.05IDD289 pKa = 3.69WGVAPIPTSTGTAPEE304 pKa = 4.02EE305 pKa = 4.53TYY307 pKa = 10.84TFADD311 pKa = 3.72SKK313 pKa = 11.02NVGMYY318 pKa = 10.08SSCEE322 pKa = 3.84NTGTAWEE329 pKa = 4.1FLKK332 pKa = 11.05FSTSEE337 pKa = 4.09EE338 pKa = 3.9QDD340 pKa = 3.14GTLLEE345 pKa = 4.66TTGQMPLRR353 pKa = 11.84ANLTDD358 pKa = 4.15AFPDD362 pKa = 3.78YY363 pKa = 10.64FATNPDD369 pKa = 3.45YY370 pKa = 8.44TTFADD375 pKa = 3.6QANRR379 pKa = 11.84TVDD382 pKa = 3.4VPNVEE387 pKa = 4.13NSVEE391 pKa = 3.97VWQTFRR397 pKa = 11.84DD398 pKa = 3.51AWTSAVIFGEE408 pKa = 4.07DD409 pKa = 4.23EE410 pKa = 3.93IPSALKK416 pKa = 10.56DD417 pKa = 3.34AAGTIDD423 pKa = 3.58EE424 pKa = 4.82LAVKK428 pKa = 10.14KK429 pKa = 10.7
Molecular weight: 45.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q5MYZ6|A0A4Q5MYZ6_9CELL GNAT family N-acetyltransferase OS=Cellulomonas sp. HLT2-17 OX=1259133 GN=EUA98_18625 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1270143 |
30 |
2576 |
335.6 |
35.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.449 ± 0.065 | 0.594 ± 0.01 |
6.078 ± 0.036 | 4.991 ± 0.038 |
2.763 ± 0.024 | 9.304 ± 0.037 |
2.007 ± 0.018 | 3.752 ± 0.03 |
1.568 ± 0.025 | 10.583 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.734 ± 0.017 | 1.682 ± 0.022 |
5.747 ± 0.037 | 2.679 ± 0.018 |
7.488 ± 0.052 | 5.33 ± 0.029 |
6.378 ± 0.04 | 9.59 ± 0.036 |
1.475 ± 0.016 | 1.808 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
