
Sorghum bicolor (Sorghum) (Sorghum vulgare)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales;
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
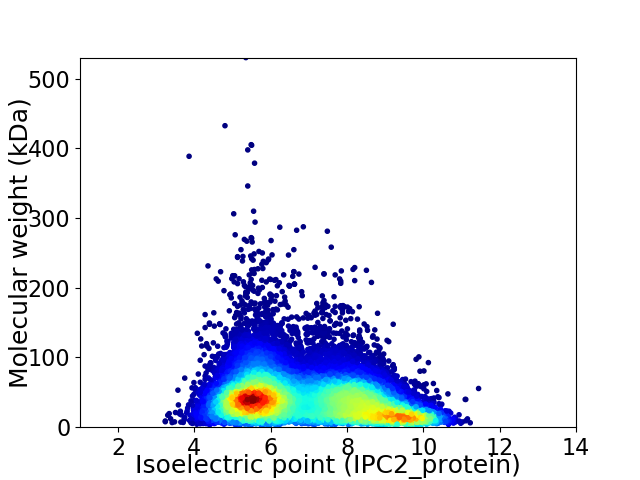
Virtual 2D-PAGE plot for 41381 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
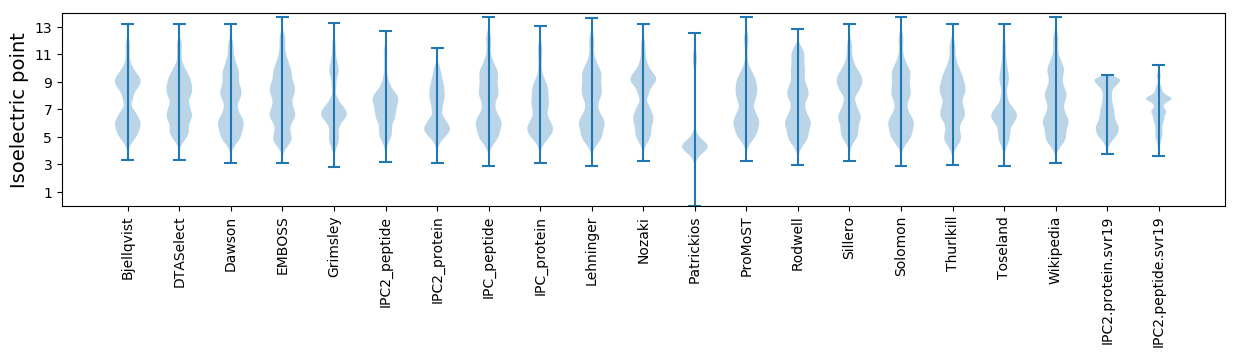
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5XAS5|C5XAS5_SORBI Uncharacterized protein OS=Sorghum bicolor OX=4558 GN=SORBI_3002G328500 PE=4 SV=1
MM1 pKa = 7.55ASSNQVLVVIVVIAVATVLAAPAAALAADD30 pKa = 4.49LLVGGSQGWRR40 pKa = 11.84LDD42 pKa = 3.31FDD44 pKa = 4.2YY45 pKa = 11.34DD46 pKa = 3.81DD47 pKa = 3.73WVEE50 pKa = 3.99EE51 pKa = 3.91NDD53 pKa = 4.65FIVGDD58 pKa = 3.85TLVFKK63 pKa = 11.08YY64 pKa = 11.01AMGQHH69 pKa = 5.59NVVQATAASYY79 pKa = 9.41AACSQGNSLQVWSSGDD95 pKa = 3.49DD96 pKa = 2.86RR97 pKa = 11.84VTLNTSGPWWFFCGVGDD114 pKa = 4.37HH115 pKa = 6.68CQDD118 pKa = 2.99GMKK121 pKa = 10.55FNINVLPAVVLSPSSPPTRR140 pKa = 11.84DD141 pKa = 2.99QGGGDD146 pKa = 3.42ATGLAGAQGGGLAAAGLAAAAGVAVVAALLFF177 pKa = 4.41
MM1 pKa = 7.55ASSNQVLVVIVVIAVATVLAAPAAALAADD30 pKa = 4.49LLVGGSQGWRR40 pKa = 11.84LDD42 pKa = 3.31FDD44 pKa = 4.2YY45 pKa = 11.34DD46 pKa = 3.81DD47 pKa = 3.73WVEE50 pKa = 3.99EE51 pKa = 3.91NDD53 pKa = 4.65FIVGDD58 pKa = 3.85TLVFKK63 pKa = 11.08YY64 pKa = 11.01AMGQHH69 pKa = 5.59NVVQATAASYY79 pKa = 9.41AACSQGNSLQVWSSGDD95 pKa = 3.49DD96 pKa = 2.86RR97 pKa = 11.84VTLNTSGPWWFFCGVGDD114 pKa = 4.37HH115 pKa = 6.68CQDD118 pKa = 2.99GMKK121 pKa = 10.55FNINVLPAVVLSPSSPPTRR140 pKa = 11.84DD141 pKa = 2.99QGGGDD146 pKa = 3.42ATGLAGAQGGGLAAAGLAAAAGVAVVAALLFF177 pKa = 4.41
Molecular weight: 17.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B6QKR8|A0A1B6QKR8_SORBI Uncharacterized protein (Fragment) OS=Sorghum bicolor OX=4558 GN=SORBI_3001G246300 PE=4 SV=1
MM1 pKa = 7.69ACTTLSLLDD10 pKa = 3.88HH11 pKa = 6.69LLITVLLILARR22 pKa = 11.84SIIIASVVVAPPPTPLPPPRR42 pKa = 11.84QRR44 pKa = 11.84APSLTPGPPPTRR56 pKa = 11.84TPLPRR61 pKa = 11.84PQLGAPARR69 pKa = 11.84GPILPPARR77 pKa = 11.84KK78 pKa = 8.94GVPPYY83 pKa = 10.59APPLPAPRR91 pKa = 11.84LGAPLPSSPLPPTRR105 pKa = 11.84APILPPRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 9.36GAPPSTPPLPTPKK127 pKa = 10.23LGAPLPSPPQPPARR141 pKa = 11.84APLLPPRR148 pKa = 11.84RR149 pKa = 11.84KK150 pKa = 10.08GPPPSTPPRR159 pKa = 11.84PAPRR163 pKa = 11.84LGVPLPSSPLPPTRR177 pKa = 11.84APVLPPRR184 pKa = 11.84RR185 pKa = 11.84KK186 pKa = 8.54GTPPSTPPRR195 pKa = 11.84PAPRR199 pKa = 11.84LGAPLPSSPLPPTRR213 pKa = 11.84APVLPPRR220 pKa = 11.84RR221 pKa = 11.84KK222 pKa = 9.4GAPPSTTPKK231 pKa = 9.38PAPRR235 pKa = 11.84LGAPFPSSPVPPTRR249 pKa = 11.84APGLPPRR256 pKa = 11.84RR257 pKa = 11.84KK258 pKa = 9.16GAPPATPPRR267 pKa = 11.84PAPRR271 pKa = 11.84LGAPLPSSPLPPTRR285 pKa = 11.84APVLPPRR292 pKa = 11.84RR293 pKa = 11.84KK294 pKa = 9.32GAPPATPPRR303 pKa = 11.84PAPRR307 pKa = 11.84LGAPLPSSPLPPTHH321 pKa = 7.04APVLPPRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.42GAPPSTPPLPTPKK343 pKa = 10.23LGAPLPSPPQPQARR357 pKa = 11.84APVLPPRR364 pKa = 11.84WKK366 pKa = 10.4GAPPSIAPQPTPRR379 pKa = 11.84LGAPLPSRR387 pKa = 11.84PQPPARR393 pKa = 11.84APLLPPRR400 pKa = 11.84QKK402 pKa = 10.54GAPPSTPPRR411 pKa = 11.84PAPRR415 pKa = 11.84LGAPLPSPILPPPARR430 pKa = 11.84ARR432 pKa = 11.84IPPPRR437 pKa = 11.84RR438 pKa = 11.84KK439 pKa = 8.63GTPPTAPRR447 pKa = 11.84FRR449 pKa = 11.84APIPSPIIPPPKK461 pKa = 9.97KK462 pKa = 10.17GIPPSTPPLPTPRR475 pKa = 11.84LGAPLPSPIIRR486 pKa = 11.84PPKK489 pKa = 10.11KK490 pKa = 9.06GTPPSIPPLPRR501 pKa = 11.84TPILPPPRR509 pKa = 11.84KK510 pKa = 9.28GAAPSSPPLRR520 pKa = 11.84PPRR523 pKa = 11.84RR524 pKa = 11.84FGAPVSSPRR533 pKa = 11.84FPPINN538 pKa = 3.43
MM1 pKa = 7.69ACTTLSLLDD10 pKa = 3.88HH11 pKa = 6.69LLITVLLILARR22 pKa = 11.84SIIIASVVVAPPPTPLPPPRR42 pKa = 11.84QRR44 pKa = 11.84APSLTPGPPPTRR56 pKa = 11.84TPLPRR61 pKa = 11.84PQLGAPARR69 pKa = 11.84GPILPPARR77 pKa = 11.84KK78 pKa = 8.94GVPPYY83 pKa = 10.59APPLPAPRR91 pKa = 11.84LGAPLPSSPLPPTRR105 pKa = 11.84APILPPRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 9.36GAPPSTPPLPTPKK127 pKa = 10.23LGAPLPSPPQPPARR141 pKa = 11.84APLLPPRR148 pKa = 11.84RR149 pKa = 11.84KK150 pKa = 10.08GPPPSTPPRR159 pKa = 11.84PAPRR163 pKa = 11.84LGVPLPSSPLPPTRR177 pKa = 11.84APVLPPRR184 pKa = 11.84RR185 pKa = 11.84KK186 pKa = 8.54GTPPSTPPRR195 pKa = 11.84PAPRR199 pKa = 11.84LGAPLPSSPLPPTRR213 pKa = 11.84APVLPPRR220 pKa = 11.84RR221 pKa = 11.84KK222 pKa = 9.4GAPPSTTPKK231 pKa = 9.38PAPRR235 pKa = 11.84LGAPFPSSPVPPTRR249 pKa = 11.84APGLPPRR256 pKa = 11.84RR257 pKa = 11.84KK258 pKa = 9.16GAPPATPPRR267 pKa = 11.84PAPRR271 pKa = 11.84LGAPLPSSPLPPTRR285 pKa = 11.84APVLPPRR292 pKa = 11.84RR293 pKa = 11.84KK294 pKa = 9.32GAPPATPPRR303 pKa = 11.84PAPRR307 pKa = 11.84LGAPLPSSPLPPTHH321 pKa = 7.04APVLPPRR328 pKa = 11.84RR329 pKa = 11.84KK330 pKa = 9.42GAPPSTPPLPTPKK343 pKa = 10.23LGAPLPSPPQPQARR357 pKa = 11.84APVLPPRR364 pKa = 11.84WKK366 pKa = 10.4GAPPSIAPQPTPRR379 pKa = 11.84LGAPLPSRR387 pKa = 11.84PQPPARR393 pKa = 11.84APLLPPRR400 pKa = 11.84QKK402 pKa = 10.54GAPPSTPPRR411 pKa = 11.84PAPRR415 pKa = 11.84LGAPLPSPILPPPARR430 pKa = 11.84ARR432 pKa = 11.84IPPPRR437 pKa = 11.84RR438 pKa = 11.84KK439 pKa = 8.63GTPPTAPRR447 pKa = 11.84FRR449 pKa = 11.84APIPSPIIPPPKK461 pKa = 9.97KK462 pKa = 10.17GIPPSTPPLPTPRR475 pKa = 11.84LGAPLPSPIIRR486 pKa = 11.84PPKK489 pKa = 10.11KK490 pKa = 9.06GTPPSIPPLPRR501 pKa = 11.84TPILPPPRR509 pKa = 11.84KK510 pKa = 9.28GAAPSSPPLRR520 pKa = 11.84PPRR523 pKa = 11.84RR524 pKa = 11.84FGAPVSSPRR533 pKa = 11.84FPPINN538 pKa = 3.43
Molecular weight: 55.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
16674116 |
29 |
5436 |
402.9 |
44.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.11 ± 0.019 | 1.995 ± 0.006 |
5.422 ± 0.009 | 5.901 ± 0.014 |
3.659 ± 0.008 | 7.202 ± 0.014 |
2.552 ± 0.006 | 4.364 ± 0.01 |
4.977 ± 0.012 | 9.527 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.005 | 3.526 ± 0.008 |
5.627 ± 0.013 | 3.563 ± 0.01 |
6.213 ± 0.011 | 8.399 ± 0.014 |
4.894 ± 0.007 | 6.766 ± 0.01 |
1.305 ± 0.004 | 2.584 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
