
Caedimonas varicaedens
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Holosporales; Caedimonadaceae; Caedimonas
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
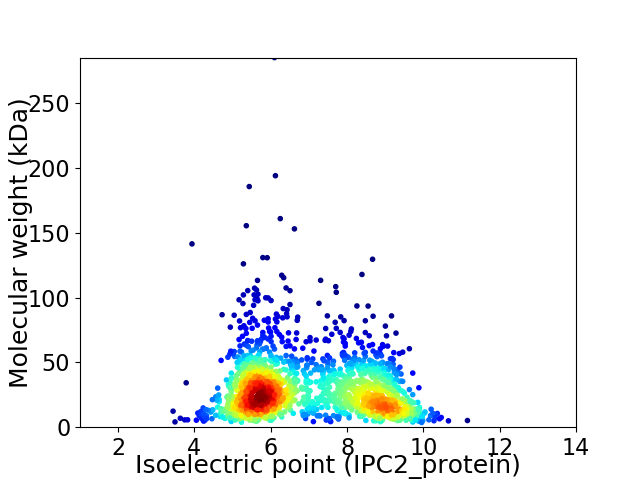
Virtual 2D-PAGE plot for 1663 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
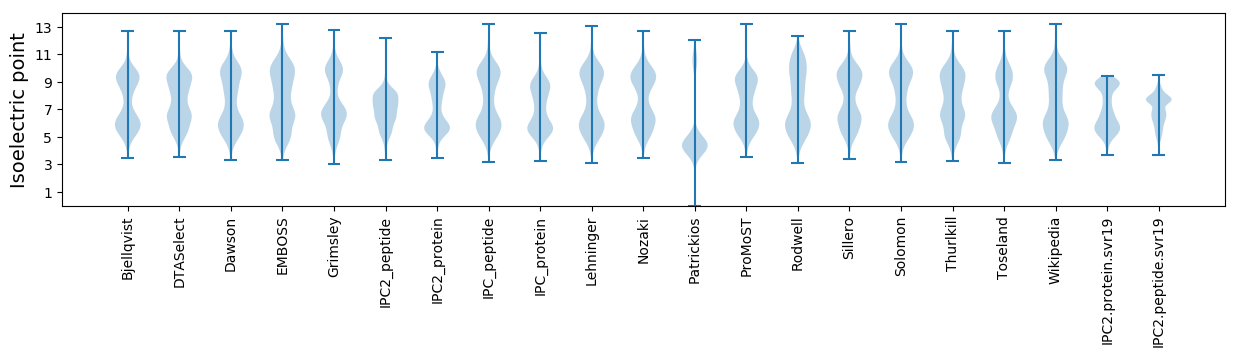
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K8MDQ4|A0A0K8MDQ4_9PROT 50S ribosomal protein L15 OS=Caedimonas varicaedens OX=1629334 GN=rplO PE=3 SV=1
MM1 pKa = 7.6LLAIDD6 pKa = 4.18ISAGGVSGDD15 pKa = 3.72NLVLQNFAVSSTADD29 pKa = 3.29PQGFYY34 pKa = 10.88AITTPGYY41 pKa = 7.32TALSIKK47 pKa = 10.71GLGTDD52 pKa = 4.03LTSGSNQLEE61 pKa = 4.27IKK63 pKa = 10.78GLDD66 pKa = 3.45THH68 pKa = 6.93AHH70 pKa = 5.13YY71 pKa = 10.83QDD73 pKa = 3.6IVDD76 pKa = 4.62NLHH79 pKa = 6.1LHH81 pKa = 6.22NNLFFTDD88 pKa = 3.76PDD90 pKa = 3.8HH91 pKa = 7.68AFGTRR96 pKa = 11.84TFSLQAFDD104 pKa = 4.86EE105 pKa = 4.97DD106 pKa = 4.3GQASATVSAPFIVTQMTAEE125 pKa = 4.08NVFTGTSGNDD135 pKa = 3.33YY136 pKa = 11.29LHH138 pKa = 6.54LTDD141 pKa = 5.79SSLTNHH147 pKa = 7.21TYY149 pKa = 10.74NAGDD153 pKa = 3.72GDD155 pKa = 5.81DD156 pKa = 4.67IIKK159 pKa = 10.73DD160 pKa = 3.62GNGPSTINGGAGHH173 pKa = 7.59DD174 pKa = 4.06LLLGQGGNDD183 pKa = 3.28ILNAGDD189 pKa = 4.93GNDD192 pKa = 3.67TLYY195 pKa = 11.26GGAGSDD201 pKa = 3.47ILIGGAGDD209 pKa = 4.3DD210 pKa = 4.15FLSGGLSGDD219 pKa = 3.71TLIGGTGKK227 pKa = 10.28DD228 pKa = 3.09VFYY231 pKa = 10.65FDD233 pKa = 4.48PSEE236 pKa = 4.37SGSLIDD242 pKa = 4.77TISDD246 pKa = 3.68FGKK249 pKa = 10.37IGSTDD254 pKa = 3.48PDD256 pKa = 4.48IIDD259 pKa = 3.12VSQYY263 pKa = 10.97LKK265 pKa = 10.76AAGYY269 pKa = 10.13IPGLNLIDD277 pKa = 3.83SFVHH281 pKa = 5.57IKK283 pKa = 9.87EE284 pKa = 4.23VSGVTTLSIDD294 pKa = 3.59PDD296 pKa = 4.02GAGPNPFTPLAVLQGIHH313 pKa = 6.34TNDD316 pKa = 3.17LVAVFDD322 pKa = 4.44GNYY325 pKa = 9.6HH326 pKa = 5.5LTVAVGG332 pKa = 3.22
MM1 pKa = 7.6LLAIDD6 pKa = 4.18ISAGGVSGDD15 pKa = 3.72NLVLQNFAVSSTADD29 pKa = 3.29PQGFYY34 pKa = 10.88AITTPGYY41 pKa = 7.32TALSIKK47 pKa = 10.71GLGTDD52 pKa = 4.03LTSGSNQLEE61 pKa = 4.27IKK63 pKa = 10.78GLDD66 pKa = 3.45THH68 pKa = 6.93AHH70 pKa = 5.13YY71 pKa = 10.83QDD73 pKa = 3.6IVDD76 pKa = 4.62NLHH79 pKa = 6.1LHH81 pKa = 6.22NNLFFTDD88 pKa = 3.76PDD90 pKa = 3.8HH91 pKa = 7.68AFGTRR96 pKa = 11.84TFSLQAFDD104 pKa = 4.86EE105 pKa = 4.97DD106 pKa = 4.3GQASATVSAPFIVTQMTAEE125 pKa = 4.08NVFTGTSGNDD135 pKa = 3.33YY136 pKa = 11.29LHH138 pKa = 6.54LTDD141 pKa = 5.79SSLTNHH147 pKa = 7.21TYY149 pKa = 10.74NAGDD153 pKa = 3.72GDD155 pKa = 5.81DD156 pKa = 4.67IIKK159 pKa = 10.73DD160 pKa = 3.62GNGPSTINGGAGHH173 pKa = 7.59DD174 pKa = 4.06LLLGQGGNDD183 pKa = 3.28ILNAGDD189 pKa = 4.93GNDD192 pKa = 3.67TLYY195 pKa = 11.26GGAGSDD201 pKa = 3.47ILIGGAGDD209 pKa = 4.3DD210 pKa = 4.15FLSGGLSGDD219 pKa = 3.71TLIGGTGKK227 pKa = 10.28DD228 pKa = 3.09VFYY231 pKa = 10.65FDD233 pKa = 4.48PSEE236 pKa = 4.37SGSLIDD242 pKa = 4.77TISDD246 pKa = 3.68FGKK249 pKa = 10.37IGSTDD254 pKa = 3.48PDD256 pKa = 4.48IIDD259 pKa = 3.12VSQYY263 pKa = 10.97LKK265 pKa = 10.76AAGYY269 pKa = 10.13IPGLNLIDD277 pKa = 3.83SFVHH281 pKa = 5.57IKK283 pKa = 9.87EE284 pKa = 4.23VSGVTTLSIDD294 pKa = 3.59PDD296 pKa = 4.02GAGPNPFTPLAVLQGIHH313 pKa = 6.34TNDD316 pKa = 3.17LVAVFDD322 pKa = 4.44GNYY325 pKa = 9.6HH326 pKa = 5.5LTVAVGG332 pKa = 3.22
Molecular weight: 34.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K8MFM7|A0A0K8MFM7_9PROT Recombination protein RecR OS=Caedimonas varicaedens OX=1629334 GN=recR PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 10.3QPSQLVRR12 pKa = 11.84KK13 pKa = 9.03RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MQTVGGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.7GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.35KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 10.3QPSQLVRR12 pKa = 11.84KK13 pKa = 9.03RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.44GFRR19 pKa = 11.84ARR21 pKa = 11.84MQTVGGRR28 pKa = 11.84RR29 pKa = 11.84VIAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.7GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
466613 |
29 |
2586 |
280.6 |
31.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.448 ± 0.072 | 1.09 ± 0.025 |
4.833 ± 0.047 | 6.442 ± 0.068 |
4.688 ± 0.059 | 6.644 ± 0.06 |
2.468 ± 0.032 | 6.983 ± 0.063 |
6.293 ± 0.059 | 10.862 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.031 | 3.681 ± 0.042 |
4.319 ± 0.046 | 4.265 ± 0.043 |
5.185 ± 0.053 | 6.633 ± 0.044 |
5.302 ± 0.038 | 6.087 ± 0.047 |
1.158 ± 0.025 | 3.103 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
