
Spathaspora sp. JA1
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Spathaspora; unclassified Spathaspora
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
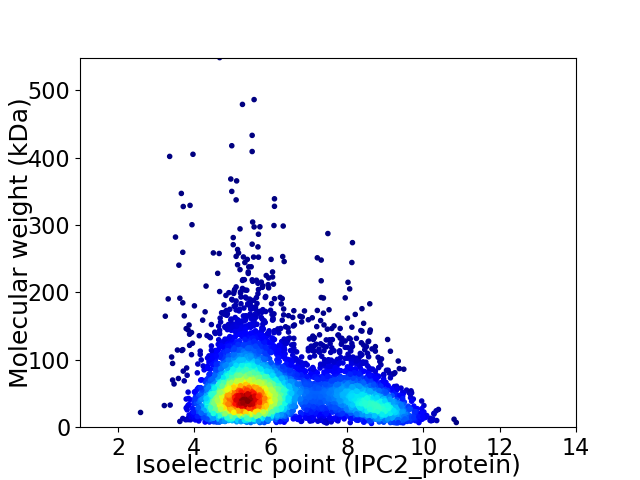
Virtual 2D-PAGE plot for 5246 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A421JR32|A0A421JR32_9ASCO Coatomer subunit alpha OS=Spathaspora sp. JA1 OX=2028339 GN=JA1_002214 PE=4 SV=1
MM1 pKa = 7.46EE2 pKa = 5.6GKK4 pKa = 10.44LEE6 pKa = 3.99EE7 pKa = 5.09DD8 pKa = 3.02KK9 pKa = 11.59LEE11 pKa = 4.24EE12 pKa = 4.14VKK14 pKa = 10.75HH15 pKa = 4.84EE16 pKa = 4.16QGHH19 pKa = 5.24KK20 pKa = 9.66VWDD23 pKa = 4.29GLDD26 pKa = 3.31HH27 pKa = 7.02SLTSNLLDD35 pKa = 4.71KK36 pKa = 9.82ITNIIEE42 pKa = 4.11CAICSEE48 pKa = 3.95IMIIPVTAEE57 pKa = 4.11CGHH60 pKa = 5.81SFCYY64 pKa = 9.99GCIYY68 pKa = 10.53QWFGTKK74 pKa = 10.19LNCPTCRR81 pKa = 11.84KK82 pKa = 10.29VIDD85 pKa = 4.46HH86 pKa = 6.75KK87 pKa = 11.02PILNIQLKK95 pKa = 9.21EE96 pKa = 3.81ISKK99 pKa = 10.7GMVDD103 pKa = 5.06LLIDD107 pKa = 3.77TDD109 pKa = 4.43SKK111 pKa = 11.86DD112 pKa = 3.45KK113 pKa = 11.18LHH115 pKa = 7.17LIGIRR120 pKa = 11.84DD121 pKa = 3.59EE122 pKa = 4.56SIKK125 pKa = 11.03SFEE128 pKa = 4.14NDD130 pKa = 3.3KK131 pKa = 10.9SCKK134 pKa = 9.99QIFGNLFKK142 pKa = 10.85GAVMLIDD149 pKa = 4.09NSDD152 pKa = 3.37GVPRR156 pKa = 11.84CGNCHH161 pKa = 5.9WEE163 pKa = 3.84AHH165 pKa = 5.99GNVCNHH171 pKa = 6.54CGTRR175 pKa = 11.84FRR177 pKa = 11.84NARR180 pKa = 11.84NMVDD184 pKa = 3.59GDD186 pKa = 3.87FDD188 pKa = 6.02GDD190 pKa = 4.26DD191 pKa = 4.13DD192 pKa = 5.73EE193 pKa = 6.29EE194 pKa = 6.0DD195 pKa = 3.63LFQDD199 pKa = 3.08VDD201 pKa = 3.99SEE203 pKa = 4.17NDD205 pKa = 3.01YY206 pKa = 11.56DD207 pKa = 5.18FEE209 pKa = 7.08DD210 pKa = 4.47GFIDD214 pKa = 3.83GRR216 pKa = 11.84TVQAIEE222 pKa = 4.44RR223 pKa = 11.84LVEE226 pKa = 4.33TYY228 pKa = 10.9GSDD231 pKa = 3.34LDD233 pKa = 5.1LYY235 pKa = 10.71EE236 pKa = 6.36DD237 pKa = 4.54PDD239 pKa = 4.05EE240 pKa = 5.13DD241 pKa = 4.99LSPSANDD248 pKa = 2.7NWFGFEE254 pKa = 4.34TDD256 pKa = 3.96DD257 pKa = 4.82SSSFEE262 pKa = 4.43LNGEE266 pKa = 4.14DD267 pKa = 5.21EE268 pKa = 5.03EE269 pKa = 6.83DD270 pKa = 3.84DD271 pKa = 4.22LSPSVIEE278 pKa = 5.13AMHH281 pKa = 6.38EE282 pKa = 3.89LHH284 pKa = 6.85NDD286 pKa = 3.49DD287 pKa = 4.23GSGSEE292 pKa = 4.21EE293 pKa = 4.68YY294 pKa = 10.75YY295 pKa = 10.75DD296 pKa = 3.56EE297 pKa = 6.68HH298 pKa = 7.47EE299 pKa = 4.63DD300 pKa = 3.33RR301 pKa = 11.84QEE303 pKa = 3.77FVEE306 pKa = 4.12EE307 pKa = 4.52HH308 pKa = 6.74EE309 pKa = 5.24DD310 pKa = 4.07GEE312 pKa = 4.9DD313 pKa = 3.36VHH315 pKa = 8.58GDD317 pKa = 3.24NDD319 pKa = 3.89GDD321 pKa = 3.72GGYY324 pKa = 9.79SEE326 pKa = 5.68DD327 pKa = 4.46HH328 pKa = 6.61NEE330 pKa = 4.26DD331 pKa = 4.27YY332 pKa = 11.18YY333 pKa = 11.63DD334 pKa = 4.1SGPGDD339 pKa = 3.47YY340 pKa = 10.7HH341 pKa = 7.51GDD343 pKa = 3.66SGQEE347 pKa = 4.07YY348 pKa = 8.96YY349 pKa = 10.93DD350 pKa = 4.07GDD352 pKa = 3.56NGYY355 pKa = 10.94DD356 pKa = 4.04DD357 pKa = 5.45GDD359 pKa = 3.47NGYY362 pKa = 10.85DD363 pKa = 4.18DD364 pKa = 5.36GGYY367 pKa = 10.66DD368 pKa = 4.41DD369 pKa = 5.98GGYY372 pKa = 10.51DD373 pKa = 5.18DD374 pKa = 4.14QEE376 pKa = 4.26SWW378 pKa = 3.2
MM1 pKa = 7.46EE2 pKa = 5.6GKK4 pKa = 10.44LEE6 pKa = 3.99EE7 pKa = 5.09DD8 pKa = 3.02KK9 pKa = 11.59LEE11 pKa = 4.24EE12 pKa = 4.14VKK14 pKa = 10.75HH15 pKa = 4.84EE16 pKa = 4.16QGHH19 pKa = 5.24KK20 pKa = 9.66VWDD23 pKa = 4.29GLDD26 pKa = 3.31HH27 pKa = 7.02SLTSNLLDD35 pKa = 4.71KK36 pKa = 9.82ITNIIEE42 pKa = 4.11CAICSEE48 pKa = 3.95IMIIPVTAEE57 pKa = 4.11CGHH60 pKa = 5.81SFCYY64 pKa = 9.99GCIYY68 pKa = 10.53QWFGTKK74 pKa = 10.19LNCPTCRR81 pKa = 11.84KK82 pKa = 10.29VIDD85 pKa = 4.46HH86 pKa = 6.75KK87 pKa = 11.02PILNIQLKK95 pKa = 9.21EE96 pKa = 3.81ISKK99 pKa = 10.7GMVDD103 pKa = 5.06LLIDD107 pKa = 3.77TDD109 pKa = 4.43SKK111 pKa = 11.86DD112 pKa = 3.45KK113 pKa = 11.18LHH115 pKa = 7.17LIGIRR120 pKa = 11.84DD121 pKa = 3.59EE122 pKa = 4.56SIKK125 pKa = 11.03SFEE128 pKa = 4.14NDD130 pKa = 3.3KK131 pKa = 10.9SCKK134 pKa = 9.99QIFGNLFKK142 pKa = 10.85GAVMLIDD149 pKa = 4.09NSDD152 pKa = 3.37GVPRR156 pKa = 11.84CGNCHH161 pKa = 5.9WEE163 pKa = 3.84AHH165 pKa = 5.99GNVCNHH171 pKa = 6.54CGTRR175 pKa = 11.84FRR177 pKa = 11.84NARR180 pKa = 11.84NMVDD184 pKa = 3.59GDD186 pKa = 3.87FDD188 pKa = 6.02GDD190 pKa = 4.26DD191 pKa = 4.13DD192 pKa = 5.73EE193 pKa = 6.29EE194 pKa = 6.0DD195 pKa = 3.63LFQDD199 pKa = 3.08VDD201 pKa = 3.99SEE203 pKa = 4.17NDD205 pKa = 3.01YY206 pKa = 11.56DD207 pKa = 5.18FEE209 pKa = 7.08DD210 pKa = 4.47GFIDD214 pKa = 3.83GRR216 pKa = 11.84TVQAIEE222 pKa = 4.44RR223 pKa = 11.84LVEE226 pKa = 4.33TYY228 pKa = 10.9GSDD231 pKa = 3.34LDD233 pKa = 5.1LYY235 pKa = 10.71EE236 pKa = 6.36DD237 pKa = 4.54PDD239 pKa = 4.05EE240 pKa = 5.13DD241 pKa = 4.99LSPSANDD248 pKa = 2.7NWFGFEE254 pKa = 4.34TDD256 pKa = 3.96DD257 pKa = 4.82SSSFEE262 pKa = 4.43LNGEE266 pKa = 4.14DD267 pKa = 5.21EE268 pKa = 5.03EE269 pKa = 6.83DD270 pKa = 3.84DD271 pKa = 4.22LSPSVIEE278 pKa = 5.13AMHH281 pKa = 6.38EE282 pKa = 3.89LHH284 pKa = 6.85NDD286 pKa = 3.49DD287 pKa = 4.23GSGSEE292 pKa = 4.21EE293 pKa = 4.68YY294 pKa = 10.75YY295 pKa = 10.75DD296 pKa = 3.56EE297 pKa = 6.68HH298 pKa = 7.47EE299 pKa = 4.63DD300 pKa = 3.33RR301 pKa = 11.84QEE303 pKa = 3.77FVEE306 pKa = 4.12EE307 pKa = 4.52HH308 pKa = 6.74EE309 pKa = 5.24DD310 pKa = 4.07GEE312 pKa = 4.9DD313 pKa = 3.36VHH315 pKa = 8.58GDD317 pKa = 3.24NDD319 pKa = 3.89GDD321 pKa = 3.72GGYY324 pKa = 9.79SEE326 pKa = 5.68DD327 pKa = 4.46HH328 pKa = 6.61NEE330 pKa = 4.26DD331 pKa = 4.27YY332 pKa = 11.18YY333 pKa = 11.63DD334 pKa = 4.1SGPGDD339 pKa = 3.47YY340 pKa = 10.7HH341 pKa = 7.51GDD343 pKa = 3.66SGQEE347 pKa = 4.07YY348 pKa = 8.96YY349 pKa = 10.93DD350 pKa = 4.07GDD352 pKa = 3.56NGYY355 pKa = 10.94DD356 pKa = 4.04DD357 pKa = 5.45GDD359 pKa = 3.47NGYY362 pKa = 10.85DD363 pKa = 4.18DD364 pKa = 5.36GGYY367 pKa = 10.66DD368 pKa = 4.41DD369 pKa = 5.98GGYY372 pKa = 10.51DD373 pKa = 5.18DD374 pKa = 4.14QEE376 pKa = 4.26SWW378 pKa = 3.2
Molecular weight: 42.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A421JM15|A0A421JM15_9ASCO Uncharacterized protein OS=Spathaspora sp. JA1 OX=2028339 GN=JA1_003636 PE=4 SV=1
MM1 pKa = 7.14YY2 pKa = 10.41KK3 pKa = 10.44VGITGKK9 pKa = 10.19FGVRR13 pKa = 11.84YY14 pKa = 9.36GSSLRR19 pKa = 11.84RR20 pKa = 11.84QTKK23 pKa = 9.43KK24 pKa = 11.15LEE26 pKa = 4.21VQQHH30 pKa = 5.25AKK32 pKa = 9.97YY33 pKa = 10.58DD34 pKa = 3.58CSFCGKK40 pKa = 8.86RR41 pKa = 11.84TVQRR45 pKa = 11.84GAAGIWNCKK54 pKa = 7.04SCRR57 pKa = 11.84KK58 pKa = 7.41TVAGGAYY65 pKa = 8.32TVSTAAAATVRR76 pKa = 11.84STIRR80 pKa = 11.84RR81 pKa = 11.84LRR83 pKa = 11.84DD84 pKa = 3.06MAEE87 pKa = 3.6AA88 pKa = 4.27
MM1 pKa = 7.14YY2 pKa = 10.41KK3 pKa = 10.44VGITGKK9 pKa = 10.19FGVRR13 pKa = 11.84YY14 pKa = 9.36GSSLRR19 pKa = 11.84RR20 pKa = 11.84QTKK23 pKa = 9.43KK24 pKa = 11.15LEE26 pKa = 4.21VQQHH30 pKa = 5.25AKK32 pKa = 9.97YY33 pKa = 10.58DD34 pKa = 3.58CSFCGKK40 pKa = 8.86RR41 pKa = 11.84TVQRR45 pKa = 11.84GAAGIWNCKK54 pKa = 7.04SCRR57 pKa = 11.84KK58 pKa = 7.41TVAGGAYY65 pKa = 8.32TVSTAAAATVRR76 pKa = 11.84STIRR80 pKa = 11.84RR81 pKa = 11.84LRR83 pKa = 11.84DD84 pKa = 3.06MAEE87 pKa = 3.6AA88 pKa = 4.27
Molecular weight: 9.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2781732 |
51 |
4819 |
530.3 |
59.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.1 ± 0.033 | 1.1 ± 0.01 |
5.788 ± 0.024 | 6.873 ± 0.04 |
4.431 ± 0.022 | 5.22 ± 0.031 |
2.102 ± 0.012 | 7.234 ± 0.029 |
6.857 ± 0.035 | 9.292 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.811 ± 0.012 | 5.843 ± 0.024 |
4.708 ± 0.036 | 4.538 ± 0.028 |
3.993 ± 0.022 | 8.594 ± 0.042 |
6.18 ± 0.047 | 5.734 ± 0.024 |
1.007 ± 0.011 | 3.594 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
